Protein Synthesis Initiation in Eukaryotic Cells
- PMID: 29735639
- PMCID: PMC6280705
- DOI: 10.1101/cshperspect.a033092
Protein Synthesis Initiation in Eukaryotic Cells
Abstract
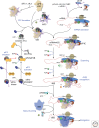
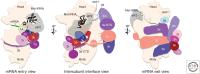
This review summarizes our current understanding of the major pathway for the initiation phase of protein synthesis in eukaryotic cells, with a focus on recent advances. We describe the major scanning or messenger RNA (mRNA) m7G cap-dependent mechanism, which is a highly coordinated and stepwise regulated process that requires the combined action of at least 12 distinct translation factors with initiator transfer RNA (tRNA), ribosomes, and mRNAs. We limit our review to studies involving either mammalian or budding yeast cells and factors, as these represent the two best-studied experimental systems, and only include a reference to other organisms where particular insight has been gained. We close with a brief description of what we feel are some of the major unknowns in eukaryotic initiation.
Copyright © 2018 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
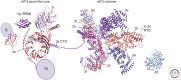

Similar articles
-
[Similar features in mechanisms of translation initiation of mRNAs in eukaryotic and prokaryotic systems].Mol Biol (Mosk). 2006 Jul-Aug;40(4):694-702. Mol Biol (Mosk). 2006. PMID: 16913228 Review. Russian.
-
Evolution and the universality of the mechanism of initiation of protein synthesis.Gene. 2009 Mar 1;432(1-2):1-6. doi: 10.1016/j.gene.2008.11.001. Epub 2008 Nov 8. Gene. 2009. PMID: 19056476 Review.
-
Four translation initiation pathways employed by the leaderless mRNA in eukaryotes.Sci Rep. 2016 Nov 28;6:37905. doi: 10.1038/srep37905. Sci Rep. 2016. PMID: 27892500 Free PMC article.
-
Eukaryotic protein synthesis.Annu Rev Biochem. 1985;54:1109-49. doi: 10.1146/annurev.bi.54.070185.005333. Annu Rev Biochem. 1985. PMID: 3896117 Review. No abstract available.
-
Initiation of translation in prokaryotes and eukaryotes.Gene. 1999 Jul 8;234(2):187-208. doi: 10.1016/s0378-1119(99)00210-3. Gene. 1999. PMID: 10395892 Review.
Cited by
-
Stress-Induced Eukaryotic Translational Regulatory Mechanisms.J Clin Med Sci. 2024;8(2):1000277. Epub 2024 Jun 24. J Clin Med Sci. 2024. PMID: 39364184 Free PMC article.
-
Ribosomal computing: implementation of the computational method.BMC Bioinformatics. 2024 Oct 3;25(1):321. doi: 10.1186/s12859-024-05945-w. BMC Bioinformatics. 2024. PMID: 39358680 Free PMC article.
-
Specific activation of the integrated stress response uncovers regulation of central carbon metabolism and lipid droplet biogenesis.Nat Commun. 2024 Sep 27;15(1):8301. doi: 10.1038/s41467-024-52538-5. Nat Commun. 2024. PMID: 39333061 Free PMC article.
-
Distinct phenotypes induced by acute hypoxia and TGF-β1 in human adult cardiac fibroblasts.J Mol Cell Cardiol Plus. 2024 Sep;9:100080. doi: 10.1016/j.jmccpl.2024.100080. Epub 2024 Jun 25. J Mol Cell Cardiol Plus. 2024. PMID: 39329164
-
Structural basis for translational control by the human 48S initiation complex.Nat Struct Mol Biol. 2024 Sep 17. doi: 10.1038/s41594-024-01378-4. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39289545
References
-
- Abramson RD, Dever TE, Lawson TG, Ray BK, Thach RE, Merrick WC. 1987. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem 262: 3826–3832. - PubMed
-
- Algire MA, Maag D, Lorsch JR. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262. - PubMed
-
- Alone PV, Dever TE. 2006. Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2Bɛ. J Biol Chem 281: 12636–12644. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
