Non-canonical activation of DAPK2 by AMPK constitutes a new pathway linking metabolic stress to autophagy
- PMID: 29717115
- PMCID: PMC5931534
- DOI: 10.1038/s41467-018-03907-4
Non-canonical activation of DAPK2 by AMPK constitutes a new pathway linking metabolic stress to autophagy
Abstract
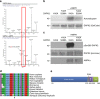
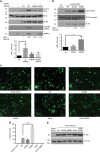
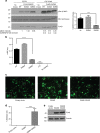
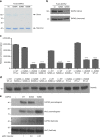
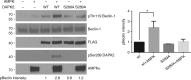
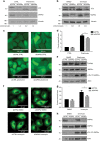
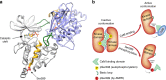
Autophagy is an intracellular degradation process essential for adaptation to metabolic stress. DAPK2 is a calmodulin-regulated protein kinase, which has been implicated in autophagy regulation, though the mechanism is unclear. Here, we show that the central metabolic sensor, AMPK, phosphorylates DAPK2 at a critical site in the protein structure, between the catalytic and the calmodulin-binding domains. This phosphorylation activates DAPK2 by functionally mimicking calmodulin binding and mitigating an inhibitory autophosphorylation, providing a novel, alternative mechanism for DAPK2 activation during metabolic stress. In addition, we show that DAPK2 phosphorylates the core autophagic machinery protein, Beclin-1, leading to dissociation of its inhibitor, Bcl-XL. Importantly, phosphorylation of DAPK2 by AMPK enhances DAPK2's ability to phosphorylate Beclin-1, and depletion of DAPK2 reduces autophagy in response to AMPK activation. Our study reveals a unique calmodulin-independent mechanism for DAPK2 activation, critical to its function as a novel downstream effector of AMPK in autophagy.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Ser289 phosphorylation activates both DAPK1 and DAPK2 but in response to different intracellular signaling pathways.Cell Cycle. 2019 Jun;18(11):1169-1176. doi: 10.1080/15384101.2019.1617616. Epub 2019 May 22. Cell Cycle. 2019. PMID: 31116076 Free PMC article.
-
DAPK2 is a novel regulator of mTORC1 activity and autophagy.Cell Death Differ. 2015 Mar;22(3):465-75. doi: 10.1038/cdd.2014.177. Epub 2014 Oct 31. Cell Death Differ. 2015. PMID: 25361081 Free PMC article.
-
Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL.Autophagy. 2009 Jul;5(5):720-2. doi: 10.4161/auto.5.5.8625. Epub 2009 Jul 2. Autophagy. 2009. PMID: 19395874
-
Death-associated protein kinase 2: Regulator of apoptosis, autophagy and inflammation.Int J Biochem Cell Biol. 2015 Aug;65:151-4. doi: 10.1016/j.biocel.2015.06.001. Epub 2015 Jun 6. Int J Biochem Cell Biol. 2015. PMID: 26055515 Review.
-
The role of DAPK2 as a key regulatory element in various human cancers: a systematic review.Mol Biol Rep. 2024 Aug 6;51(1):886. doi: 10.1007/s11033-024-09761-6. Mol Biol Rep. 2024. PMID: 39105958 Review.
Cited by
-
Crosstalk between let-7a-5p and BCL-xL in the Initiation of Toxic Autophagy in Lung Cancer.Mol Ther Oncolytics. 2019 Sep 10;15:69-78. doi: 10.1016/j.omto.2019.08.010. eCollection 2019 Dec 20. Mol Ther Oncolytics. 2019. PMID: 31650027 Free PMC article.
-
Using Monozygotic Twins to Dissect Common Genes in Posttraumatic Stress Disorder and Migraine.Front Neurosci. 2021 Jun 22;15:678350. doi: 10.3389/fnins.2021.678350. eCollection 2021. Front Neurosci. 2021. PMID: 34239411 Free PMC article.
-
Genome-Wide Atlas of Promoter Expression Reveals Contribution of Transcribed Regulatory Elements to Genetic Control of Disuse-Mediated Atrophy of Skeletal Muscle.Biology (Basel). 2021 Jun 20;10(6):557. doi: 10.3390/biology10060557. Biology (Basel). 2021. PMID: 34203013 Free PMC article.
-
BECLIN1: Protein Structure, Function and Regulation.Cells. 2021 Jun 17;10(6):1522. doi: 10.3390/cells10061522. Cells. 2021. PMID: 34204202 Free PMC article. Review.
-
miR-1285-3p Controls Colorectal Cancer Proliferation and Escape from Apoptosis through DAPK2.Int J Mol Sci. 2020 Mar 31;21(7):2423. doi: 10.3390/ijms21072423. Int J Mol Sci. 2020. PMID: 32244500 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

