Emergence of the Asian lineage dengue virus type 3 genotype III in Malaysia
- PMID: 29699483
- PMCID: PMC5921268
- DOI: 10.1186/s12862-018-1175-4
Emergence of the Asian lineage dengue virus type 3 genotype III in Malaysia
Abstract
Background: Dengue virus type 3 genotype III (DENV3/III) is associated with increased number of severe infections when it emerged in the Americas and Asia. We had previously demonstrated that the DENV3/III was introduced into Malaysia in the late 2000s. We investigated the genetic diversity of DENV3/III strains recovered from Malaysia and examined their phylogenetic relationships against other DENV3/III strains isolated globally.
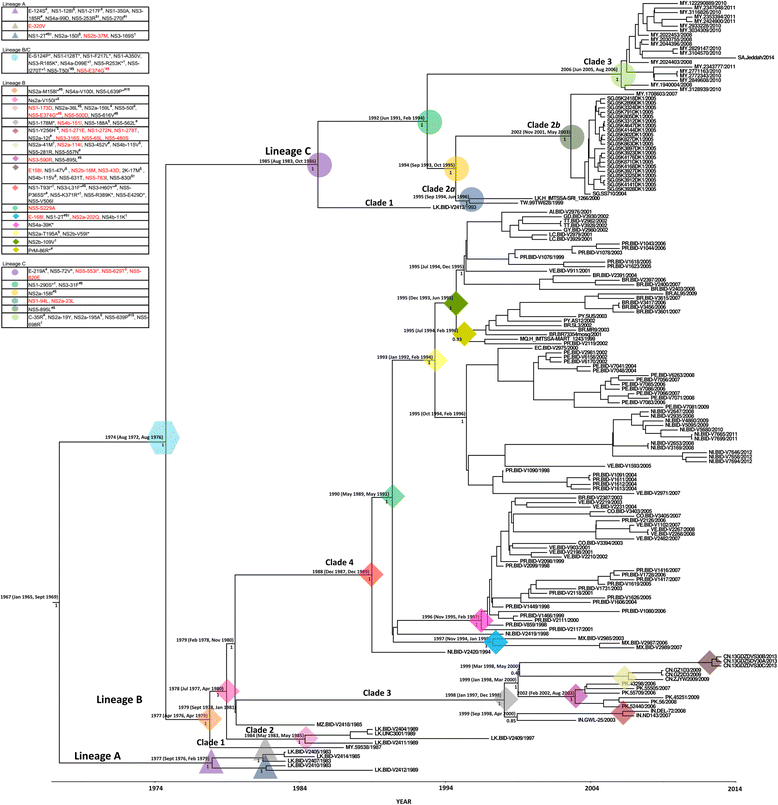
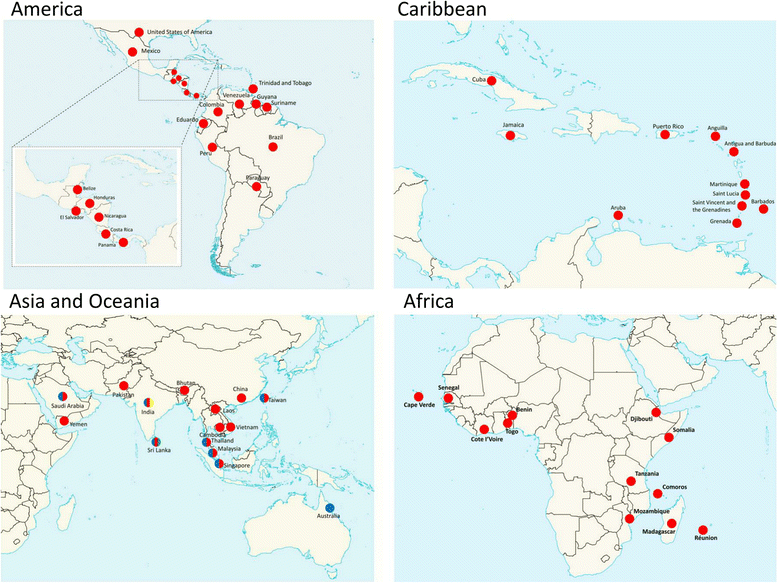
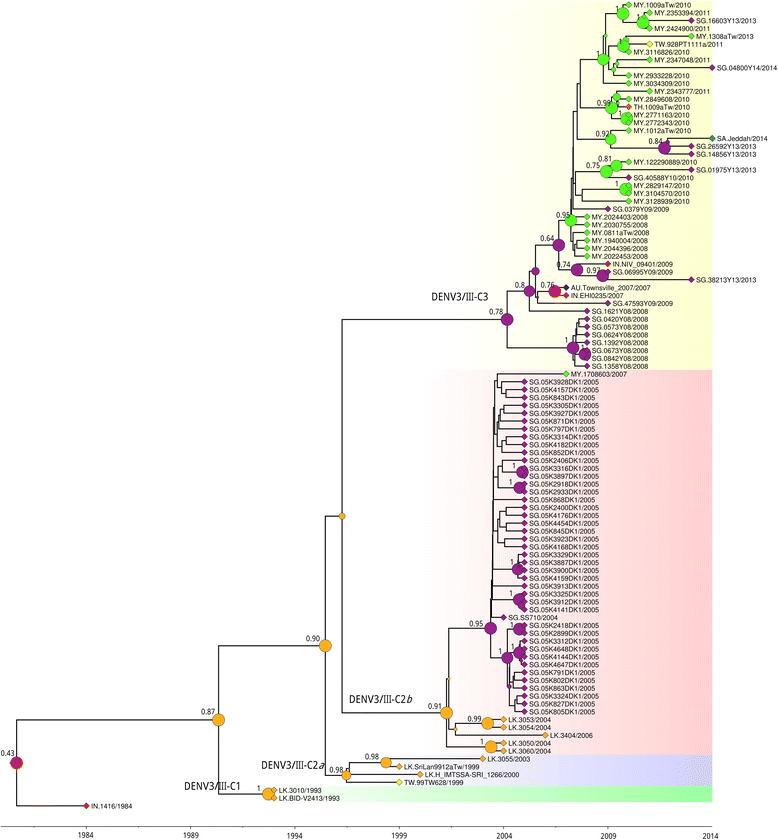
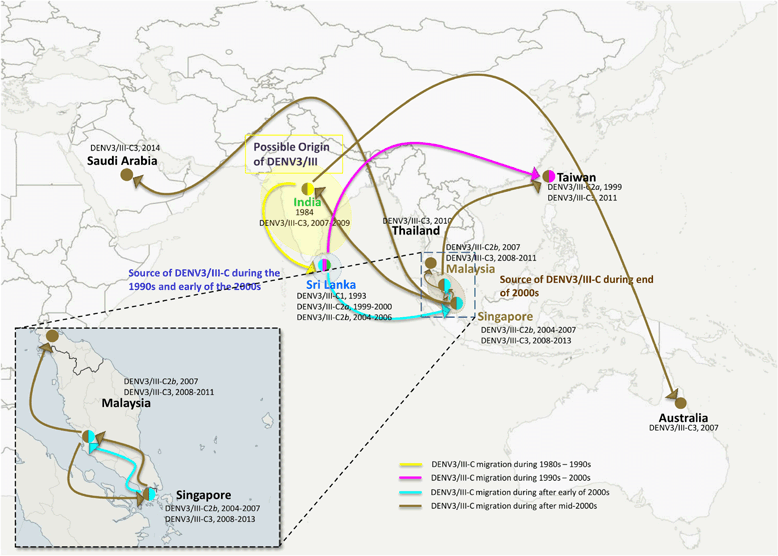
Results: Phylogenetic analysis revealed at least four distinct DENV3/III lineages. Two of the lineages (DENV3/III-B and DENV3/III-C) are current actively circulating whereas the DENV3/III-A and DENV3/III-D were no longer recovered since the 1980s. Selection pressure analysis revealed strong evidence of positive selection on a number of amino acid sites in PrM, E, NS1, NS2a, NS2b, NS3, NS4a, and NS5. The Malaysian DENV3/III isolates recovered in the 1980s (MY.59538/1987) clustered into DENV3/III-B, which was the lineage with cosmopolitan distribution consisting of strains actively circulating in the Americas, Africa, and Asia. The Malaysian isolates recovered after the 2000s clustered within DENV3/III-C. This DENV3/III-C lineage displayed a more restricted geographical distribution and consisted of isolates recovered from Asia, denoted as the Asian lineage. Amino acid variation sites in NS5 (NS5-553I/M, NS5-629 T, and NS5-820E) differentiated the DENV3/III-C from other DENV3 viruses. The codon 629 of NS5 was identified as a positively selected site. While the NS5-698R was identified as unique to the genome of DENV3/III-C3. Phylogeographic results suggested that the recent Malaysian DENV3/III-C was likely to have been introduced from Singapore in 2008 and became endemic. From Malaysia, the virus subsequently spread into Taiwan and Thailand in the early part of the 2010s and later reintroduced into Singapore in 2013.
Conclusions: Distinct clustering of the Malaysian old and new DENV3/III isolates suggests that the currently circulating DENV3/III in Malaysia did not descend directly from the strains recovered during the 1980s. Phylogenetic analyses and common genetic traits in the genome of the strains and those from the neighboring countries suggest that the Malaysian DENV3/III is likely to have been introduced from the neighboring regions. Malaysia, however, serves as one of the sources of the recent regional spread of DENV3/III-C3 within the Asia region.
Keywords: Arbovirus; DENV3, phylogenetic; Dengue virus; Infectious disease; Malaysia.
Conflict of interest statement
Authors’ information
SAB is a senior professor and Director of the Tropical Infectious Diseases Research and Education Centre (TIDREC) at University of Malaya. He is also the director the WHO Collaborating Centre for Arbovirus Reference & Research (Dengue/Severe Dengue) at the University of Malaya. His research focus is on emerging infectious diseases particularly vector-borne and zoonotic diseases in the tropics. KKT is a Ph.D. candidate in the Department of Medical Microbiology and researcher with the Tropical Infectious Diseases Research and Education Centre (TIDREC) at University of Malaya. Her research interest is in evolutionary genomics of infectious diseases.
Ethics approval and consent to participate
The use of the data and virus isolates in this study were approved by the Medical Ethic Committee of the UMMC with MEC Ref No of 806.23 and 806.24.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Disruption of predicted dengue virus type 3 major outbreak cycle coincided with switching of the dominant circulating virus genotype.Infect Genet Evol. 2017 Oct;54:271-275. doi: 10.1016/j.meegid.2017.07.008. Epub 2017 Jul 8. Infect Genet Evol. 2017. PMID: 28698156
-
The Complete Genome Phylogeny of Geographically Distinct Dengue Virus Serotype 2 Isolates (1944-2013) Supports Further Groupings within the Cosmopolitan Genotype.PLoS One. 2015 Sep 28;10(9):e0138900. doi: 10.1371/journal.pone.0138900. eCollection 2015. PLoS One. 2015. PMID: 26414178 Free PMC article.
-
Phylogenetic analysis of dengue virus types 1 and 3 isolated in Jakarta, Indonesia in 1988.Infect Genet Evol. 2012 Dec;12(8):1938-43. doi: 10.1016/j.meegid.2012.08.006. Epub 2012 Aug 31. Infect Genet Evol. 2012. PMID: 22959957
-
The origin, emergence and evolutionary genetics of dengue virus.Infect Genet Evol. 2003 May;3(1):19-28. doi: 10.1016/s1567-1348(03)00004-2. Infect Genet Evol. 2003. PMID: 12797969 Review.
-
Epidemiology of dengue disease in Malaysia (2000-2012): a systematic literature review.PLoS Negl Trop Dis. 2014 Nov 6;8(11):e3159. doi: 10.1371/journal.pntd.0003159. eCollection 2014. PLoS Negl Trop Dis. 2014. PMID: 25375211 Free PMC article. Review.
Cited by
-
Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand.PLoS One. 2018 Nov 12;13(11):e0207220. doi: 10.1371/journal.pone.0207220. eCollection 2018. PLoS One. 2018. PMID: 30419004 Free PMC article.
-
Differential heterologous neutralisation profile against strains within DENV-3 genotype II.Epidemiol Infect. 2021 Dec 17;150:e33. doi: 10.1017/S0950268821002648. Epidemiol Infect. 2021. PMID: 35225194 Free PMC article.
-
Re-Emergence of DENV-3 in French Guiana: Retrospective Analysis of Cases That Circulated in the French Territories of the Americas from the 2000s to the 2023-2024 Outbreak.Viruses. 2024 Aug 14;16(8):1298. doi: 10.3390/v16081298. Viruses. 2024. PMID: 39205272 Free PMC article.
-
Long-Term Protection Elicited by a DNA Vaccine Candidate Expressing the prM-E Antigen of Dengue Virus Serotype 3 in Mice.Front Cell Infect Microbiol. 2020 Mar 17;10:87. doi: 10.3389/fcimb.2020.00087. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32257963 Free PMC article.
-
Re-Emergence of Dengue Serotype 3 in the Context of a Large Religious Gathering Event in Touba, Senegal.Int J Environ Res Public Health. 2022 Dec 16;19(24):16912. doi: 10.3390/ijerph192416912. Int J Environ Res Public Health. 2022. PMID: 36554793 Free PMC article.
References
-
- Prince HE, Yeh C, Lape-Nixon M. Primary and probable secondary dengue virus (DV) infection rates in relation to age among DV IgM-positive patients residing in the United States mainland versus the Caribbean islands. Clin Vaccine Immunol. 2012;19(1):105–108. doi: 10.1128/CVI.05519-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

