Cross-sectional analysis of CD8 T cell immunity to human herpesvirus 6B
- PMID: 29698478
- PMCID: PMC5919459
- DOI: 10.1371/journal.ppat.1006991
Cross-sectional analysis of CD8 T cell immunity to human herpesvirus 6B
Abstract
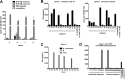
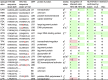
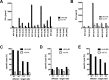
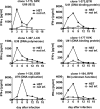
Human herpesvirus 6 (HHV-6) is prevalent in healthy persons, causes disease in immunosuppressed carriers, and may be involved in autoimmune disease. Cytotoxic CD8 T cells are probably important for effective control of infection. However, the HHV-6-specific CD8 T cell repertoire is largely uncharacterized. Therefore, we undertook a virus-wide analysis of CD8 T cell responses to HHV-6. We used a simple anchor motif-based algorithm (SAMBA) to identify 299 epitope candidates potentially presented by the HLA class I molecule B*08:01. Candidates were found in 77 of 98 unique HHV-6B proteins. From peptide-expanded T cell lines, we obtained CD8 T cell clones against 20 candidates. We tested whether T cell clones recognized HHV-6-infected cells. This was the case for 16 epitopes derived from 12 proteins from all phases of the viral replication cycle. Epitopes were enriched in certain amino acids flanking the peptide. Ex vivo analysis of eight healthy donors with HLA-peptide multimers showed that the strongest responses were directed against an epitope from IE-2, with a median frequency of 0.09% of CD8 T cells. Reconstitution of T cells specific for this and other HHV-6 epitopes was also observed after allogeneic hematopoietic stem cell transplantation. We conclude that HHV-6 induces CD8 T cell responses against multiple antigens of diverse functional classes. Most antigens against which CD8 T cells can be raised are presented by infected cells. Ex vivo multimer staining can directly identify HHV-6-specific T cells. These results will advance development of immune monitoring, adoptive T cell therapy, and vaccines.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Specific CD8⁺ T cells recognize human herpesvirus 6B.Eur J Immunol. 2012 Nov;42(11):2901-12. doi: 10.1002/eji.201242439. Epub 2012 Sep 26. Eur J Immunol. 2012. PMID: 22886850
-
Human herpesvirus 6B immediate-early I protein contains functional HLA-A*02, HLA-A*03, and HLA-B*07 class I restricted CD8(+) T-cell epitopes.Eur J Immunol. 2014 Dec;44(12):3573-84. doi: 10.1002/eji.201444931. Epub 2014 Oct 29. Eur J Immunol. 2014. PMID: 25243920 Clinical Trial.
-
Genome-Wide Approach to the CD4 T-Cell Response to Human Herpesvirus 6B.J Virol. 2019 Jun 28;93(14):e00321-19. doi: 10.1128/JVI.00321-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31043533 Free PMC article.
-
Immune response to HHV-6 and implications for immunotherapy.Curr Opin Virol. 2014 Dec;9:154-61. doi: 10.1016/j.coviro.2014.10.001. Epub 2014 Oct 27. Curr Opin Virol. 2014. PMID: 25462448 Free PMC article. Review.
-
HHV-6 and the immune system: mechanisms of immunomodulation and viral escape.J Clin Virol. 2006 Dec;37 Suppl 1:S4-10. doi: 10.1016/S1386-6532(06)70004-X. J Clin Virol. 2006. PMID: 17276368 Review.
Cited by
-
Immunoinformatic Analysis Reveals Antigenic Heterogeneity of Epstein-Barr Virus Is Immune-Driven.Front Immunol. 2021 Dec 16;12:796379. doi: 10.3389/fimmu.2021.796379. eCollection 2021. Front Immunol. 2021. PMID: 34975903 Free PMC article.
-
Potent high-avidity neutralizing antibodies and T cell responses after COVID-19 vaccination in individuals with B cell lymphoma and multiple myeloma.Nat Cancer. 2023 Jan;4(1):81-95. doi: 10.1038/s43018-022-00502-x. Epub 2022 Dec 21. Nat Cancer. 2023. PMID: 36543907 Free PMC article.
-
Advances in the Characterization of the T-Cell Response to Human Herpesvirus-6.Front Immunol. 2018 Jun 25;9:1454. doi: 10.3389/fimmu.2018.01454. eCollection 2018. Front Immunol. 2018. PMID: 29988505 Free PMC article. Review.
-
Evasion of the Host Immune Response by Betaherpesviruses.Int J Mol Sci. 2021 Jul 13;22(14):7503. doi: 10.3390/ijms22147503. Int J Mol Sci. 2021. PMID: 34299120 Free PMC article. Review.
-
Phenotypic and Functional Differences between Human Herpesvirus 6- and Human Cytomegalovirus-Specific T Cells.J Virol. 2019 Jun 14;93(13):e02321-18. doi: 10.1128/JVI.02321-18. Print 2019 Jul 1. J Virol. 2019. PMID: 30996090 Free PMC article.
References
-
- Baillargeon J, Piper J, Leach CT (2000) Epidemiology of human herpesvirus 6 (HHV-6) infection in pregnant and nonpregnant women. J Clin Virol 16: 149–157. - PubMed
-
- De Bolle L, Naesens L, De Clercq E (2005) Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev 18: 217–245. doi: 10.1128/CMR.18.1.217-245.2005 - DOI - PMC - PubMed
-
- Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y et al. (1988) Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet 1: 1065–1067. - PubMed
-
- Zerr DM, Meier AS, Selke SS, Frenkel LM, Huang ML, Wald A et al. (2005) A population-based study of primary human herpesvirus 6 infection. N Engl J Med 352: 768–776. doi: 10.1056/NEJMoa042207 - DOI - PubMed
-
- Agut H, Bonnafous P, Gautheret-Dejean A (2015) Laboratory and clinical aspects of human herpesvirus 6 infections. Clin Microbiol Rev 28: 313–335. doi: 10.1128/CMR.00122-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

