Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity
- PMID: 29691590
- PMCID: PMC11105402
- DOI: 10.1007/s00018-018-2820-1
Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity
Abstract
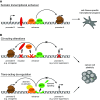
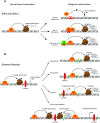
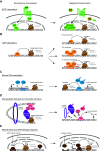
Cancer heterogeneity arises during tumor progression as a consequence of genetic insults, environmental cues, and reversible changes in the epigenetic state, favoring tumor cell plasticity. The role of enhancer reprogramming is emerging as a relevant field in cancer biology as it supports adaptation of cancer cells to those environmental changes encountered during tumor progression and metastasis seeding. In this review, we describe the cancer-related alterations that drive oncogenic enhancer activity, leading to dysregulated transcriptional programs. We discuss the molecular mechanisms of both cis- and trans-factors in overriding the regulatory circuits that maintain cell-type specificity and imposing an alternative, de-regulated enhancer activity in cancer cells. We further comment on the increasing evidence which implicates stress response and aging-signaling pathways in the enhancer landscape reprogramming during tumorigenesis. Finally, we focus on the potential therapeutic implications of these enhancer-mediated subverted transcriptional programs, putting particular emphasis on the lack of information regarding tumor progression and the metastatic outgrowth, which still remain the major cause of mortality related to cancer.
Keywords: Cancer; Cis-regulatory elements; DNA damage; Enhancer; Epigenetic; Metastasis; Reprogramming; Signaling pathways; Transcription factors; Tumor progression.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures
Similar articles
-
Mathematical models of cell phenotype regulation and reprogramming: Make cancer cells sensitive again!Biochim Biophys Acta Rev Cancer. 2017 Apr;1867(2):167-175. doi: 10.1016/j.bbcan.2017.04.001. Epub 2017 Apr 7. Biochim Biophys Acta Rev Cancer. 2017. PMID: 28396217 Free PMC article. Review.
-
Epigenetic plasticity of enhancers in cancer.Transcription. 2020 Feb;11(1):26-36. doi: 10.1080/21541264.2020.1713682. Epub 2020 Jan 16. Transcription. 2020. PMID: 31944157 Free PMC article. Review.
-
Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity.Trends Cancer. 2017 May;3(5):372-386. doi: 10.1016/j.trecan.2017.04.004. Epub 2017 May 5. Trends Cancer. 2017. PMID: 28718414 Free PMC article. Review.
-
Context-Dependent Epigenetic Regulation of Nuclear Factor of Activated T Cells 1 in Pancreatic Plasticity.Gastroenterology. 2017 May;152(6):1507-1520.e15. doi: 10.1053/j.gastro.2017.01.043. Epub 2017 Feb 8. Gastroenterology. 2017. PMID: 28188746
-
Metabolic reprogramming directed by super-enhancers in tumors: An emerging landscape.Mol Ther. 2024 Mar 6;32(3):572-579. doi: 10.1016/j.ymthe.2024.02.003. Epub 2024 Feb 6. Mol Ther. 2024. PMID: 38327048 Review.
Cited by
-
Transcriptome profiling reveals that VNPP433-3β, the lead next-generation galeterone analog inhibits prostate cancer stem cells by downregulating epithelial-mesenchymal transition and stem cell markers.Mol Carcinog. 2022 Jul;61(7):643-654. doi: 10.1002/mc.23406. Epub 2022 May 5. Mol Carcinog. 2022. PMID: 35512605 Free PMC article.
-
Potential application of cell reprogramming techniques for cancer research.Cell Mol Life Sci. 2019 Jan;76(1):45-65. doi: 10.1007/s00018-018-2924-7. Epub 2018 Oct 3. Cell Mol Life Sci. 2019. PMID: 30283976 Free PMC article. Review.
-
Cancer cell plasticity, stem cell factors, and therapy resistance: how are they linked?Cancer Metastasis Rev. 2024 Mar;43(1):423-440. doi: 10.1007/s10555-023-10144-9. Epub 2023 Oct 5. Cancer Metastasis Rev. 2024. PMID: 37796391 Review.
-
Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness.Cancers (Basel). 2020 Dec 11;12(12):3716. doi: 10.3390/cancers12123716. Cancers (Basel). 2020. PMID: 33322354 Free PMC article. Review.
-
MUC1-C Activates the NuRD Complex to Drive Dedifferentiation of Triple-Negative Breast Cancer Cells.Cancer Res. 2019 Nov 15;79(22):5711-5722. doi: 10.1158/0008-5472.CAN-19-1034. Epub 2019 Sep 13. Cancer Res. 2019. PMID: 31519689 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

