Hypoxia and Chromatin: A Focus on Transcriptional Repression Mechanisms
- PMID: 29690561
- PMCID: PMC6027312
- DOI: 10.3390/biomedicines6020047
Hypoxia and Chromatin: A Focus on Transcriptional Repression Mechanisms
Abstract
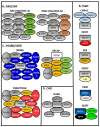
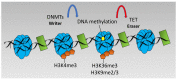
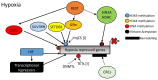
Hypoxia or reduced oxygen availability has been studied extensively for its ability to activate specific genes. Hypoxia-induced gene expression is mediated by the HIF transcription factors, but not exclusively so. Despite the extensive knowledge about how hypoxia activates genes, much less is known about how hypoxia promotes gene repression. In this review, we discuss the potential mechanisms underlying hypoxia-induced transcriptional repression responses. We highlight HIF-dependent and independent mechanisms as well as the potential roles of dioxygenases with functions at the nucleosome and DNA level. Lastly, we discuss recent evidence regarding the involvement of transcriptional repressor complexes in hypoxia.
Keywords: HIF; JmjC; chromatin; histone methylation; hypoxia; repressor Complexes; transcriptional repression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
JmjC histone demethylases act as chromatin oxygen sensors.Mol Cell Oncol. 2019 May 7;6(4):1608501. doi: 10.1080/23723556.2019.1608501. eCollection 2019. Mol Cell Oncol. 2019. PMID: 31211238 Free PMC article.
-
Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast.Mol Cell Biol. 2000 Oct;20(19):7088-98. doi: 10.1128/MCB.20.19.7088-7098.2000. Mol Cell Biol. 2000. PMID: 10982825 Free PMC article.
-
Inhibition of the Oxygen-Sensing Asparaginyl Hydroxylase Factor Inhibiting Hypoxia-Inducible Factor: A Potential Hypoxia Response Modulating Strategy.J Med Chem. 2021 Jun 10;64(11):7189-7209. doi: 10.1021/acs.jmedchem.1c00415. Epub 2021 May 24. J Med Chem. 2021. PMID: 34029087 Review.
-
Transcriptional repression of hypoxia-inducible factor-1 (HIF-1) by the protein arginine methyltransferase PRMT1.Mol Biol Cell. 2014 Mar;25(6):925-35. doi: 10.1091/mbc.E13-07-0423. Epub 2014 Jan 22. Mol Biol Cell. 2014. PMID: 24451260 Free PMC article.
-
Hypoxia-driven epigenetic regulation in cancer progression: A focus on histone methylation and its modifying enzymes.Cancer Lett. 2020 Oct 1;489:41-49. doi: 10.1016/j.canlet.2020.05.025. Epub 2020 Jun 6. Cancer Lett. 2020. PMID: 32522693 Review.
Cited by
-
Hypoxia and aging: molecular mechanisms, diseases, and therapeutic targets.MedComm (2020). 2024 Oct 15;5(11):e786. doi: 10.1002/mco2.786. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39415849 Free PMC article. Review.
-
Hypoxic reactivation of Kaposi's sarcoma associated herpesvirus.Cell Insight. 2024 Sep 7;3(6):100200. doi: 10.1016/j.cellin.2024.100200. eCollection 2024 Dec. Cell Insight. 2024. PMID: 39391006 Free PMC article. Review.
-
Hypoxia in the Blue Mussel Mytilus chilensis Induces a Transcriptome Shift Associated with Endoplasmic Reticulum Stress, Metabolism, and Immune Response.Genes (Basel). 2024 May 22;15(6):658. doi: 10.3390/genes15060658. Genes (Basel). 2024. PMID: 38927594 Free PMC article.
-
Transcriptional Responses of Different Brain Cell Types to Oxygen Decline.Brain Sci. 2024 Mar 30;14(4):341. doi: 10.3390/brainsci14040341. Brain Sci. 2024. PMID: 38671993 Free PMC article.
-
Nrf2 sensitizes ferroptosis through l-2-hydroxyglutarate-mediated chromatin modifications in sickle cell disease.Blood. 2023 Jul 27;142(4):382-396. doi: 10.1182/blood.2022018159. Blood. 2023. PMID: 37267508 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

