Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity
- PMID: 29684204
- PMCID: PMC5961234
- DOI: 10.1093/nar/gky268
Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity
Abstract
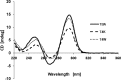
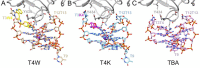
Thrombin-binding aptamer (TBA) is a DNA 15-mer of sequence 5'-GGT TGG TGT GGT TGG-3' that folds into a G-quadruplex structure linked by two T-T loops located on one side and a T-G-T loop on the other. These loops are critical for post-SELEX modification to improve TBA target affinity. With this goal in mind we synthesized a T analog, 5-(indolyl-3-acetyl-3-amino-1-propenyl)-2'-deoxyuridine (W) to substitute one T or a pair of Ts. Subsequently, the affinity for each analog was determined by biolayer interferometry. An aptamer with W at position 4 exhibited about 3-fold increased binding affinity, and replacing both T4 and T12 with W afforded an almost 10-fold enhancement compared to native TBA. To better understand the role of the substituent's aromatic moiety, an aptamer with 5-(methyl-3-acetyl-3-amino-1-propenyl)-2'-deoxyuridine (K; W without the indole moiety) in place of T4 was also synthesized. This K4 aptamer was found to improve affinity 7-fold relative to native TBA. Crystal structures of aptamers with T4 replaced by either W or K bound to thrombin provide insight into the origins of the increased affinities. Our work demonstrates that facile chemical modification of a simple DNA aptamer can be used to significantly improve its binding affinity for a well-established pharmacological target protein.
Figures
Similar articles
-
Impact of the Position of the Chemically Modified 5-Furyl-2'-Deoxyuridine Nucleoside on the Thrombin DNA Aptamer-Protein Complex: Structural Insights into Aptamer Response from MD Simulations.Molecules. 2019 Aug 10;24(16):2908. doi: 10.3390/molecules24162908. Molecules. 2019. PMID: 31405145 Free PMC article.
-
Structural and Binding Effects of Chemical Modifications on Thrombin Binding Aptamer (TBA).Molecules. 2021 Jul 30;26(15):4620. doi: 10.3390/molecules26154620. Molecules. 2021. PMID: 34361773 Free PMC article.
-
Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer.FEBS J. 2013 Dec;280(24):6581-8. doi: 10.1111/febs.12561. Epub 2013 Nov 1. FEBS J. 2013. PMID: 24128303
-
Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications.Curr Pharm Des. 2012;18(14):2036-47. doi: 10.2174/138161212799958387. Curr Pharm Des. 2012. PMID: 22376107 Review.
-
G-quadruplex-based aptamers targeting human thrombin: Discovery, chemical modifications and antithrombotic effects.Pharmacol Ther. 2021 Jan;217:107649. doi: 10.1016/j.pharmthera.2020.107649. Epub 2020 Aug 7. Pharmacol Ther. 2021. PMID: 32777331 Review.
Cited by
-
Impact of the Position of the Chemically Modified 5-Furyl-2'-Deoxyuridine Nucleoside on the Thrombin DNA Aptamer-Protein Complex: Structural Insights into Aptamer Response from MD Simulations.Molecules. 2019 Aug 10;24(16):2908. doi: 10.3390/molecules24162908. Molecules. 2019. PMID: 31405145 Free PMC article.
-
Hybrid-Type SELEX for the Selection of Artificial Nucleic Acid Aptamers Exhibiting Cell Internalization Activity.Pharmaceutics. 2021 Jun 15;13(6):888. doi: 10.3390/pharmaceutics13060888. Pharmaceutics. 2021. PMID: 34204006 Free PMC article.
-
Structural and Binding Effects of Chemical Modifications on Thrombin Binding Aptamer (TBA).Molecules. 2021 Jul 30;26(15):4620. doi: 10.3390/molecules26154620. Molecules. 2021. PMID: 34361773 Free PMC article.
-
Aptamers for Proteins Associated with Rheumatic Diseases: Progress, Challenges, and Prospects of Diagnostic and Therapeutic Applications.Biomedicines. 2020 Nov 22;8(11):527. doi: 10.3390/biomedicines8110527. Biomedicines. 2020. PMID: 33266394 Free PMC article. Review.
-
Structural Biology for the Molecular Insight between Aptamers and Target Proteins.Int J Mol Sci. 2021 Apr 15;22(8):4093. doi: 10.3390/ijms22084093. Int J Mol Sci. 2021. PMID: 33920991 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

