IFN-λ and microRNAs are important modulators of the pulmonary innate immune response against influenza A (H1N2) infection in pigs
- PMID: 29677213
- PMCID: PMC5909910
- DOI: 10.1371/journal.pone.0194765
IFN-λ and microRNAs are important modulators of the pulmonary innate immune response against influenza A (H1N2) infection in pigs
Abstract
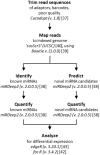
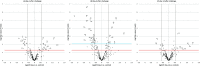
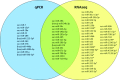
The innate immune system is paramount in the response to and clearance of influenza A virus (IAV) infection in non-immune individuals. Known factors include type I and III interferons and antiviral pathogen recognition receptors, and the cascades of antiviral and pro- and anti-inflammatory gene expression they induce. MicroRNAs (miRNAs) are increasingly recognized to participate in post-transcriptional modulation of these responses, but the temporal dynamics of how these players of the antiviral innate immune response collaborate to combat infection remain poorly characterized. We quantified the expression of miRNAs and protein coding genes in the lungs of pigs 1, 3, and 14 days after challenge with swine IAV (H1N2). Through RT-qPCR we observed a 400-fold relative increase in IFN-λ3 gene expression on day 1 after challenge, and a strong interferon-mediated antiviral response was observed on days 1 and 3 accompanied by up-regulation of genes related to the pro-inflammatory response and apoptosis. Using small RNA sequencing and qPCR validation we found 27 miRNAs that were differentially expressed after challenge, with the highest number of regulated miRNAs observed on day 3. In contrast, the number of protein coding genes found to be regulated due to IAV infection peaked on day 1. Pulmonary miRNAs may thus be aimed at fine-tuning the initial rapid inflammatory response after IAV infection. Specifically, we found five miRNAs (ssc-miR-15a, ssc-miR-18a, ssc-miR-21, ssc-miR-29b, and hsa-miR-590-3p)-four known porcine miRNAs and one novel porcine miRNA candidate-to be potential modulators of viral pathogen recognition and apoptosis. A total of 11 miRNAs remained differentially expressed 14 days after challenge, at which point the infection had cleared. In conclusion, the results suggested a role for miRNAs both during acute infection as well as later, with the potential to influence lung homeostasis and susceptibility to secondary infections in the lungs of pigs after IAV infection.
Conflict of interest statement
Figures
Similar articles
-
Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2).Innate Immun. 2013 Oct;19(5):531-44. doi: 10.1177/1753425912473668. Epub 2013 Feb 12. Innate Immun. 2013. PMID: 23405029
-
Pulmonary MicroRNA expression after heterologous challenge with swine influenza A virus (H1N2) in immunized and non-immunized pigs.Virology. 2024 Aug;596:110117. doi: 10.1016/j.virol.2024.110117. Epub 2024 May 23. Virology. 2024. PMID: 38797064
-
Late regulation of immune genes and microRNAs in circulating leukocytes in a pig model of influenza A (H1N2) infection.Sci Rep. 2016 Feb 19;6:21812. doi: 10.1038/srep21812. Sci Rep. 2016. PMID: 26893019 Free PMC article.
-
Porcine innate and adaptative immune responses to influenza and coronavirus infections.Ann N Y Acad Sci. 2006 Oct;1081(1):130-6. doi: 10.1196/annals.1373.014. Ann N Y Acad Sci. 2006. PMID: 17135502 Free PMC article. Review.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
Cited by
-
miRNA Regulatory Functions in Farm Animal Diseases, and Biomarker Potentials for Effective Therapies.Int J Mol Sci. 2021 Mar 17;22(6):3080. doi: 10.3390/ijms22063080. Int J Mol Sci. 2021. PMID: 33802936 Free PMC article. Review.
-
Prognostic significance of serum miR-18a-5p in severe COVID-19 Egyptian patients.J Genet Eng Biotechnol. 2023 Nov 13;21(1):114. doi: 10.1186/s43141-023-00565-y. J Genet Eng Biotechnol. 2023. PMID: 37953403 Free PMC article.
-
MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock.Front Vet Sci. 2020 Dec 18;7:578193. doi: 10.3389/fvets.2020.578193. eCollection 2020. Front Vet Sci. 2020. PMID: 33392281 Free PMC article. Review.
-
Innate antiviral responses in porcine nasal mucosal explants inoculated with influenza A virus are comparable with responses in respiratory tissues after viral infection.Immunobiology. 2022 May;227(3):152192. doi: 10.1016/j.imbio.2022.152192. Epub 2022 Feb 22. Immunobiology. 2022. PMID: 35255458 Free PMC article.
-
Upregulation of miR-101 during Influenza A Virus Infection Abrogates Viral Life Cycle by Targeting mTOR Pathway.Viruses. 2020 Apr 15;12(4):444. doi: 10.3390/v12040444. Viruses. 2020. PMID: 32326380 Free PMC article.
References
-
- Mauskopf J, Klesse M, Lee S, Herrera-Taracena G. The burden of influenza complications in different high-risk groups: a targeted literature review. J. Med. Econ. 2013;16:264–77. doi: 10.3111/13696998.2012.752376 - DOI - PubMed
-
- Wiersma L, Rimmelzwaan G, de Vries R. Developing Universal Influenza Vaccines: Hitting the Nail, Not Just on the Head. Vaccines. 2015;3:239–62. doi: 10.3390/vaccines3020239 - DOI - PMC - PubMed
-
- Tripathi S, White MR, Hartshorn KL. The amazing innate immune response to influenza A virus infection. Innate Immun. 2013;21:73–98. doi: 10.1177/1753425913508992 - DOI - PubMed
-
- Kotenko S V, Gallagher G, Baurin V V, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

