Endosomal receptor trafficking: Retromer and beyond
- PMID: 29667289
- PMCID: PMC6043395
- DOI: 10.1111/tra.12574
Endosomal receptor trafficking: Retromer and beyond
Abstract
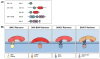
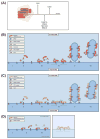
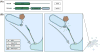
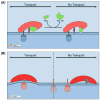
The tubular endolysosomal network is a quality control system that ensures the proper delivery of internalized receptors to specific subcellular destinations in order to maintain cellular homeostasis. Although retromer was originally described in yeast as a regulator of endosome-to-Golgi receptor recycling, mammalian retromer has emerged as a central player in endosome-to-plasma membrane recycling of a variety of receptors. Over the past decade, information regarding the mechanism by which retromer facilitates receptor trafficking has emerged, as has the identification of numerous retromer-associated molecules including the WASH complex, sorting nexins (SNXs) and TBC1d5. Moreover, the recent demonstration that several SNXs can directly interact with retromer cargo to facilitate endosome-to-Golgi retrieval has provided new insight into how these receptors are trafficked in cells. The mechanism by which SNX17 cargoes are recycled out of the endosomal system was demonstrated to involve a retromer-like complex termed the retriever, which is recruited to WASH positive endosomes through an interaction with the COMMD/CCDC22/CCDC93 (CCC) complex. Lastly, the mechanisms by which bacterial and viral pathogens highjack this complex sorting machinery in order to escape the endolysosomal system or remain hidden within the cells are beginning to emerge. In this review, we will highlight recent studies that have begun to unravel the intricacies by which the retromer and associated molecules contribute to receptor trafficking and how deregulation at this sorting domain can contribute to disease or facilitate pathogen infection.
Keywords: WASH; endosome; receptor trafficking; retriever; retromer; sorting nexin.
© 2018 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures

Similar articles
-
Towards a molecular understanding of endosomal trafficking by Retromer and Retriever.Traffic. 2019 Jul;20(7):465-478. doi: 10.1111/tra.12649. Traffic. 2019. PMID: 30993794 Review.
-
Receptor Recycling by Retromer.Mol Cell Biol. 2023;43(7):317-334. doi: 10.1080/10985549.2023.2222053. Epub 2023 Jun 23. Mol Cell Biol. 2023. PMID: 37350516 Free PMC article. Review.
-
FAM21 directs SNX27-retromer cargoes to the plasma membrane by preventing transport to the Golgi apparatus.Nat Commun. 2016 Mar 9;7:10939. doi: 10.1038/ncomms10939. Nat Commun. 2016. PMID: 26956659 Free PMC article.
-
The retromer complex - endosomal protein recycling and beyond.J Cell Sci. 2012 Oct 15;125(Pt 20):4693-702. doi: 10.1242/jcs.103440. Epub 2012 Nov 12. J Cell Sci. 2012. PMID: 23148298 Free PMC article. Review.
-
Retromer and sorting nexins in endosomal sorting.Biochem Soc Trans. 2015 Feb;43(1):33-47. doi: 10.1042/BST20140290. Biochem Soc Trans. 2015. PMID: 25619244 Review.
Cited by
-
Implication of repeat insertion domains in the trans-activity of the long non-coding RNA ANRIL.Nucleic Acids Res. 2021 May 21;49(9):4954-4970. doi: 10.1093/nar/gkab245. Nucleic Acids Res. 2021. PMID: 33872355 Free PMC article.
-
The adaptor protein PID1 regulates receptor-dependent endocytosis of postprandial triglyceride-rich lipoproteins.Mol Metab. 2018 Oct;16:88-99. doi: 10.1016/j.molmet.2018.07.010. Epub 2018 Jul 30. Mol Metab. 2018. PMID: 30100244 Free PMC article.
-
Endosome Traffic Modulates Pro-Inflammatory Signal Transduction in CD4+ T Cells-Implications for the Pathogenesis of Systemic Lupus Erythematosus.Int J Mol Sci. 2023 Jun 28;24(13):10749. doi: 10.3390/ijms241310749. Int J Mol Sci. 2023. PMID: 37445926 Free PMC article. Review.
-
Metabolic regulation through the endosomal system.Traffic. 2019 Aug;20(8):552-570. doi: 10.1111/tra.12670. Epub 2019 Jun 24. Traffic. 2019. PMID: 31177593 Free PMC article. Review.
-
Retromer dependent changes in cellular homeostasis and Parkinson's disease.Essays Biochem. 2021 Dec 22;65(7):987-998. doi: 10.1042/EBC20210023. Essays Biochem. 2021. PMID: 34528672 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

