Identification of Unequally Represented Founder Viruses Among Tissues in Very Early SIV Rectal Transmission
- PMID: 29651274
- PMCID: PMC5884942
- DOI: 10.3389/fmicb.2018.00557
Identification of Unequally Represented Founder Viruses Among Tissues in Very Early SIV Rectal Transmission
Abstract
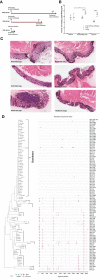
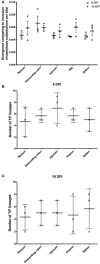
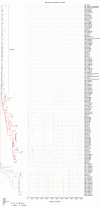
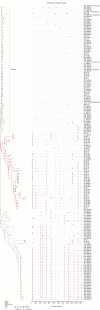
Characterizing the transmitted/founder (T/F) viruses of multi-variant SIV infection may shed new light on the understanding of mucosal transmission. We intrarectally inoculated six Chinese rhesus macaques with a single high dose of SIVmac251 (3.1 × 104 TCID50) and obtained 985 full-length env sequences from multiple tissues at 6 and 10 days post-infection by single genome amplification (SGA). All 6 monkeys were infected with a range of 2 to 8 T/F viruses and the dominant variants from the inoculum were still dominant in different tissues from each monkey. Interestingly, our data showed that a cluster of rare T/F viruses was unequally represented in different tissues. This cluster of rare T/F viruses phylogenetically related to the non-dominant SIV variants in the inoculum and was not detected in any rectum tissues, but could be identified in the descending colon, jejunum, spleen, or plasma. In 2 out of 6 macaques, identical SIVmac251 variants belonging to this cluster were detected simultaneously in descending colon/jejunum and the inoculum. We also demonstrated that the average CG dinucleotide frequency of these rare T/F viruses found in tissues, as well as non-dominant variants in the inoculum, was significantly higher than the dominant T/F viruses in tissues and the inoculum. Collectively, these findings suggest that descending colon/jejunum might be more susceptible than rectum to SIV in the very early phase of infection. And host CG suppression, which was previously shown to inhibit HIV replication in vitro, may also contribute to the bottleneck selection during in vivo transmission.
Keywords: CG dinucleotide; Chinese rhesus macaque; SIV; rectal transmission; transmitted/founder virus; very early virological events; zinc finger antiviral protein.
Figures

Similar articles
-
Characterization of founder viruses in very early SIV rectal transmission.Virology. 2017 Feb;502:97-105. doi: 10.1016/j.virol.2016.12.018. Epub 2016 Dec 25. Virology. 2017. PMID: 28027479 Free PMC article.
-
Conserved molecular signatures in gp120 are associated with the genetic bottleneck during simian immunodeficiency virus (SIV), SIV-human immunodeficiency virus (SHIV), and HIV type 1 (HIV-1) transmission.J Virol. 2015 Apr;89(7):3619-29. doi: 10.1128/JVI.03235-14. Epub 2015 Jan 14. J Virol. 2015. PMID: 25589663 Free PMC article.
-
Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques.J Virol. 2001 Apr;75(8):3753-65. doi: 10.1128/JVI.75.8.3753-3765.2001. J Virol. 2001. PMID: 11264364 Free PMC article.
-
Tracking the Emergence of Host-Specific Simian Immunodeficiency Virus env and nef Populations Reveals nef Early Adaptation and Convergent Evolution in Brain of Naturally Progressing Rhesus Macaques.J Virol. 2015 Aug;89(16):8484-96. doi: 10.1128/JVI.01010-15. Epub 2015 Jun 3. J Virol. 2015. PMID: 26041280 Free PMC article.
-
Persistent Viral Reservoirs in Lymphoid Tissues in SIV-Infected Rhesus Macaques of Chinese-Origin on Suppressive Antiretroviral Therapy.Viruses. 2019 Jan 27;11(2):105. doi: 10.3390/v11020105. Viruses. 2019. PMID: 30691203 Free PMC article.
Cited by
-
European Non-Polio Enterovirus Network: Introduction of Hospital-Based Surveillance Network to Understand the True Disease Burden of Non-Polio Enterovirus and Parechovirus Infections in Europe.Microorganisms. 2021 Aug 27;9(9):1827. doi: 10.3390/microorganisms9091827. Microorganisms. 2021. PMID: 34576722 Free PMC article.
References
-
- Cline A. N., Bess J. W., Piatak M. J., Lifson J. D. (2005). Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34, 303–312. 10.1111/j.1600-0684.2005.00128.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

