Major Determinants of Nucleosome Positioning
- PMID: 29628211
- PMCID: PMC6129461
- DOI: 10.1016/j.bpj.2018.03.015
Major Determinants of Nucleosome Positioning
Abstract
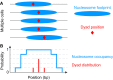
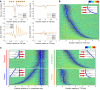
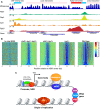
The compact structure of the nucleosome limits DNA accessibility and inhibits the binding of most sequence-specific proteins. Nucleosomes are not randomly located on the DNA but positioned with respect to the DNA sequence, suggesting models in which critical binding sites are either exposed in the linker, resulting in activation, or buried inside a nucleosome, resulting in repression. The mechanisms determining nucleosome positioning are therefore of paramount importance for understanding gene regulation and other events that occur in chromatin, such as transcription, replication, and repair. Here, we review our current understanding of the major determinants of nucleosome positioning: DNA sequence, nonhistone DNA-binding proteins, chromatin-remodeling enzymes, and transcription. We outline the major challenges for the future: elucidating the precise mechanisms of chromatin opening and promoter activation, identifying the complexes that occupy promoters, and understanding the multiscale problem of chromatin fiber organization.
Published by Elsevier Inc.
Figures

Similar articles
-
Chromatin associated mechanisms in base excision repair - nucleosome remodeling and DNA transcription, two key players.Free Radic Biol Med. 2017 Jun;107:159-169. doi: 10.1016/j.freeradbiomed.2016.12.026. Epub 2016 Dec 20. Free Radic Biol Med. 2017. PMID: 28011149 Review.
-
Nucleosome dynamics during chromatin remodeling in vivo.Nucleus. 2016;7(1):20-6. doi: 10.1080/19491034.2016.1149666. Epub 2016 Mar 2. Nucleus. 2016. PMID: 26933790 Free PMC article.
-
Nucleosome positioning in relation to nucleosome spacing and DNA sequence-specific binding of a protein.FEBS J. 2007 May;274(9):2396-410. doi: 10.1111/j.1742-4658.2007.05775.x. Epub 2007 Apr 5. FEBS J. 2007. PMID: 17419736
-
Sequence-specific targeting of chromatin remodelers organizes precisely positioned nucleosomes throughout the genome.Bioessays. 2017 Jan;39(1):1-8. doi: 10.1002/bies.201600183. Epub 2016 Nov 16. Bioessays. 2017. PMID: 27862071 Free PMC article. Review.
-
ATP-dependent nucleosome remodeling.Annu Rev Biochem. 2002;71:247-73. doi: 10.1146/annurev.biochem.71.110601.135400. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045097 Review.
Cited by
-
CORENup: a combination of convolutional and recurrent deep neural networks for nucleosome positioning identification.BMC Bioinformatics. 2020 Sep 16;21(Suppl 8):326. doi: 10.1186/s12859-020-03627-x. BMC Bioinformatics. 2020. PMID: 32938377 Free PMC article.
-
Good News for Nuclear Transgene Expression in Chlamydomonas.Cells. 2019 Nov 28;8(12):1534. doi: 10.3390/cells8121534. Cells. 2019. PMID: 31795196 Free PMC article. Review.
-
Chromatin replication and epigenetic cell memory.Nat Cell Biol. 2020 Apr;22(4):361-371. doi: 10.1038/s41556-020-0487-y. Epub 2020 Mar 30. Nat Cell Biol. 2020. PMID: 32231312 Review.
-
Accessibility of promoter DNA is not the primary determinant of chromatin-mediated gene regulation.Genome Res. 2019 Dec;29(12):1985-1995. doi: 10.1101/gr.249326.119. Epub 2019 Sep 11. Genome Res. 2019. PMID: 31511305 Free PMC article.
-
Dynamic nucleosome remodeling mediated by YY1 underlies early mouse development.Genes Dev. 2023 Jul 1;37(13-14):590-604. doi: 10.1101/gad.350376.122. Epub 2023 Aug 2. Genes Dev. 2023. PMID: 37532472 Free PMC article.
References
-
- Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- van Holde K.E. Springer; New York: 1989. Chromatin.
-
- Clark D.J., Kimura T. Electrostatic mechanism of chromatin folding. J. Mol. Biol. 1990;211:883–896. - PubMed
-
- Felsenfeld G., Clark D., Studitsky V. Transcription through nucleosomes. Biophys. Chem. 2000;86:231–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

