Transcriptional Regulation of the Peripheral Pathway for the Anaerobic Catabolism of Toluene and m-Xylene in Azoarcus sp. CIB
- PMID: 29623071
- PMCID: PMC5874301
- DOI: 10.3389/fmicb.2018.00506
Transcriptional Regulation of the Peripheral Pathway for the Anaerobic Catabolism of Toluene and m-Xylene in Azoarcus sp. CIB
Abstract
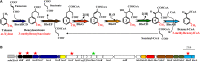
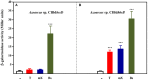
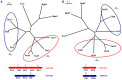
Alkylbenzenes, such as toluene and m-xylene, are an important class of contaminant hydrocarbons that are widespread and tend to accumulate in subsurface anoxic environments. The peripheral pathway for the anaerobic oxidation of toluene in bacteria consists of an initial activation catalyzed by a benzylsuccinate synthase (encoded by bss genes), and a subsequent modified β-oxidation of benzylsuccinate to benzoyl-CoA and succinyl-CoA (encoded by bbs genes). We have shown here that the bss and bbs genes, which are located within an integrative and conjugative element, are essential for anaerobic degradation of toluene but also for m-xylene oxidation in the denitrifying beta-proteobacterium Azoarcus sp. CIB. New insights into the transcriptional organization and regulation of a complete gene cluster for anaerobic catabolism of toluene/m-xylene in a single bacterial strain are presented. The bss and bbs genes are transcriptionally coupled into two large convergent catabolic operons driven by the PbssD and PbbsA promoters, respectively, whose expression is inducible when cells grow anaerobically in toluene or m-xylene. An adjacent tdiSR operon driven by the PtdiS promoter encodes a putative two-component regulatory system. TdiR behaves as a transcriptional activator of the PbssD, PbbsA, and PtdiS promoters, being benzylsuccinate/(3-methyl)benzylsuccinate, rather than toluene/m-xylene, the inducers that may trigger the TdiS-mediated activation of TdiR. In addition to the TdiSR-based specific control, the expression of the bss and bbs genes in Azoarcus sp. CIB is under an overimposed regulation that depends on certain environmental factors, such as the presence/absence of oxygen or the availability of preferred carbon sources (catabolite repression). This work paves the way for future strategies toward the reliable assessment of microbial activity in toluene/m-xylene contaminated environments.
Keywords: Azoarcus; anaerobic degradation; biomarker; catabolite repression; m-xylene; tdiSR; toluene.
Figures


Similar articles
-
Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization.J Bacteriol. 2001 Dec;183(23):6763-70. doi: 10.1128/JB.183.23.6763-6770.2001. J Bacteriol. 2001. PMID: 11698363 Free PMC article.
-
Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1.Arch Microbiol. 2004 Mar;181(3):182-94. doi: 10.1007/s00203-003-0627-3. Epub 2004 Jan 21. Arch Microbiol. 2004. PMID: 14735297
-
The ICEXTD of Azoarcus sp. CIB, an integrative and conjugative element with aerobic and anaerobic catabolic properties.Environ Microbiol. 2016 Dec;18(12):5018-5031. doi: 10.1111/1462-2920.13465. Epub 2016 Aug 30. Environ Microbiol. 2016. PMID: 27450529
-
Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria.Biodegradation. 2000;11(2-3):85-105. doi: 10.1023/a:1011122631799. Biodegradation. 2000. PMID: 11440245 Review.
-
Microbial aspects of anaerobic BTEX degradation.Biomed Environ Sci. 2002 Jun;15(2):130-44. Biomed Environ Sci. 2002. PMID: 12244754 Review.
Cited by
-
Enhancing the Rice Seedlings Growth Promotion Abilities of Azoarcus sp. CIB by Heterologous Expression of ACC Deaminase to Improve Performance of Plants Exposed to Cadmium Stress.Microorganisms. 2020 Sep 22;8(9):1453. doi: 10.3390/microorganisms8091453. Microorganisms. 2020. PMID: 32971998 Free PMC article.
-
ArxA From Azoarcus sp. CIB, an Anaerobic Arsenite Oxidase From an Obligate Heterotrophic and Mesophilic Bacterium.Front Microbiol. 2019 Jul 30;10:1699. doi: 10.3389/fmicb.2019.01699. eCollection 2019. Front Microbiol. 2019. PMID: 31417512 Free PMC article.
-
Golden Standard: a complete standard, portable, and interoperative MoClo tool for model and non-model proteobacteria.Nucleic Acids Res. 2023 Oct 27;51(19):e98. doi: 10.1093/nar/gkad758. Nucleic Acids Res. 2023. PMID: 37718823 Free PMC article.
-
Enhancing tellurite and selenite bioconversions by overexpressing a methyltransferase from Aromatoleum sp. CIB.Microb Biotechnol. 2023 May;16(5):915-930. doi: 10.1111/1751-7915.14162. Epub 2022 Nov 10. Microb Biotechnol. 2023. PMID: 36366868 Free PMC article.
-
The impact of calcium peroxide on groundwater bacterial diversity during naphthalene removal by permeable reactive barrier (PRB).Environ Sci Pollut Res Int. 2019 Dec;26(34):35218-35226. doi: 10.1007/s11356-019-06398-y. Epub 2019 Nov 6. Environ Sci Pollut Res Int. 2019. PMID: 31691896
References
-
- Achong G. R., Rodriguez A. M., Spormann A. M. (2001). Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183 6763–6770. 10.1128/JB.183.23.6763-6770.2001 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

