Gene transfers can date the tree of life
- PMID: 29610471
- PMCID: PMC5912509
- DOI: 10.1038/s41559-018-0525-3
Gene transfers can date the tree of life
Abstract
Biodiversity has always been predominantly microbial, and the scarcity of fossils from bacteria, archaea and microbial eukaryotes has prevented a comprehensive dating of the tree of life. Here, we show that patterns of lateral gene transfer deduced from an analysis of modern genomes encode a novel and abundant source of information about the temporal coexistence of lineages throughout the history of life. We use state-of-the-art species tree-aware phylogenetic methods to reconstruct the history of thousands of gene families and demonstrate that dates implied by gene transfers are consistent with estimates from relaxed molecular clocks in Bacteria, Archaea and Eukarya. We present the order of speciations according to lateral gene transfer data calibrated to geological time for three datasets comprising 40 genomes for Cyanobacteria, 60 genomes for Archaea and 60 genomes for Fungi. An inspection of discrepancies between transfers and clocks and a comparison with mammalian fossils show that gene transfer in microbes is potentially as informative for dating the tree of life as the geological record in macroorganisms.
Conflict of interest statement
The authors declare no competing interests.
Figures
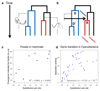

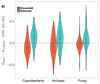

Comment in
-
Fossil-free dating.Nat Ecol Evol. 2018 May;2(5):771-772. doi: 10.1038/s41559-018-0532-4. Nat Ecol Evol. 2018. PMID: 29610470 No abstract available.
Similar articles
-
Relative Time Constraints Improve Molecular Dating.Syst Biol. 2022 Jun 16;71(4):797-809. doi: 10.1093/sysbio/syab084. Syst Biol. 2022. PMID: 34668564 Free PMC article.
-
Genome-scale phylogenetic analysis finds extensive gene transfer among fungi.Philos Trans R Soc Lond B Biol Sci. 2015 Sep 26;370(1678):20140335. doi: 10.1098/rstb.2014.0335. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26323765 Free PMC article.
-
Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations.Proc Natl Acad Sci U S A. 2012 Oct 23;109(43):17513-8. doi: 10.1073/pnas.1202997109. Epub 2012 Oct 4. Proc Natl Acad Sci U S A. 2012. PMID: 23043116 Free PMC article.
-
En route to a genome-based classification of Archaea and Bacteria?Syst Appl Microbiol. 2010 Jun;33(4):175-82. doi: 10.1016/j.syapm.2010.03.003. Epub 2010 Apr 20. Syst Appl Microbiol. 2010. PMID: 20409658 Review.
-
Genome trees and the nature of genome evolution.Annu Rev Microbiol. 2005;59:191-209. doi: 10.1146/annurev.micro.59.030804.121233. Annu Rev Microbiol. 2005. PMID: 16153168 Review.
Cited by
-
Features of a novel protein, rusticalin, from the ascidian Styela rustica reveal ancestral horizontal gene transfer event.Mob DNA. 2019 Jan 19;10:4. doi: 10.1186/s13100-019-0146-7. eCollection 2019. Mob DNA. 2019. PMID: 30675192 Free PMC article.
-
Phylogenetic reconciliation: making the most of genomes to understand microbial ecology and evolution.ISME J. 2024 Jan 8;18(1):wrae129. doi: 10.1093/ismejo/wrae129. ISME J. 2024. PMID: 39001714 Free PMC article. Review.
-
Transposon clusters as substrates for aberrant splice-site activation.RNA Biol. 2021 Mar;18(3):354-367. doi: 10.1080/15476286.2020.1805909. Epub 2020 Sep 23. RNA Biol. 2021. PMID: 32965162 Free PMC article.
-
Zombi: a phylogenetic simulator of trees, genomes and sequences that accounts for dead linages.Bioinformatics. 2020 Feb 15;36(4):1286-1288. doi: 10.1093/bioinformatics/btz710. Bioinformatics. 2020. PMID: 31566657 Free PMC article.
-
Reconciling multiple genes trees via segmental duplications and losses.Algorithms Mol Biol. 2019 Mar 20;14:7. doi: 10.1186/s13015-019-0139-6. eCollection 2019. Algorithms Mol Biol. 2019. PMID: 30930955 Free PMC article.
References
-
- Zuckerkandl E, Pauling . Molecular Disease, Evolution, and Genic Heterogeneity. 1962.
-
- O’Leary MA, et al. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science. 2013;339:662–667. - PubMed
-
- Donoghue PCJ, Benton MJ. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol Evol. 2007;22:424–431. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

