Signaling Pathways Involved in the Regulation of mRNA Translation
- PMID: 29610153
- PMCID: PMC5974435
- DOI: 10.1128/MCB.00070-18
Signaling Pathways Involved in the Regulation of mRNA Translation
Abstract
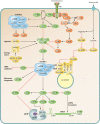
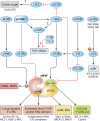
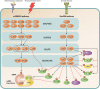
Translation is a key step in the regulation of gene expression and one of the most energy-consuming processes in the cell. In response to various stimuli, multiple signaling pathways converge on the translational machinery to regulate its function. To date, the roles of phosphoinositide 3-kinase (PI3K)/AKT and the mitogen-activated protein kinase (MAPK) pathways in the regulation of translation are among the best understood. Both pathways engage the mechanistic target of rapamycin (mTOR) to regulate a variety of components of the translational machinery. While these pathways regulate protein synthesis in homeostasis, their dysregulation results in aberrant translation leading to human diseases, including diabetes, neurological disorders, and cancer. Here we review the roles of the PI3K/AKT and MAPK pathways in the regulation of mRNA translation. We also highlight additional signaling mechanisms that have recently emerged as regulators of the translational apparatus.
Keywords: MAPK; MNK; RSK; eIF4E; mRNA; mRNA translation; mTOR; mitogen-activated protein kinases; protein phosphorylation; signal transduction; translational control.
Copyright © 2018 Roux and Topisirovic.
Figures
Similar articles
-
Regulation of mRNA translation by signaling pathways.Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a012252. doi: 10.1101/cshperspect.a012252. Cold Spring Harb Perspect Biol. 2012. PMID: 22888049 Free PMC article. Review.
-
Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells.Carcinogenesis. 2008 Dec;29(12):2279-88. doi: 10.1093/carcin/bgn221. Epub 2008 Sep 22. Carcinogenesis. 2008. PMID: 18809972
-
Translational control by oncogenic signaling pathways.Biochim Biophys Acta. 2015 Jul;1849(7):753-65. doi: 10.1016/j.bbagrm.2014.11.006. Epub 2014 Dec 2. Biochim Biophys Acta. 2015. PMID: 25477072 Review.
-
Newcastle Disease virus infection activates PI3K/Akt/mTOR and p38 MAPK/Mnk1 pathways to benefit viral mRNA translation via interaction of the viral NP protein and host eIF4E.PLoS Pathog. 2020 Jun 30;16(6):e1008610. doi: 10.1371/journal.ppat.1008610. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32603377 Free PMC article.
-
Increased milk protein synthesis in response to exogenous growth hormone is associated with changes in mechanistic (mammalian) target of rapamycin (mTOR)C1-dependent and independent cell signaling.J Dairy Sci. 2013 Apr;96(4):2327-2338. doi: 10.3168/jds.2012-6267. Epub 2013 Feb 22. J Dairy Sci. 2013. PMID: 23462168
Cited by
-
eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS.Acta Neuropathol Commun. 2019 Apr 25;7(1):62. doi: 10.1186/s40478-019-0711-9. Acta Neuropathol Commun. 2019. PMID: 31023341 Free PMC article.
-
Leishmania donovani Lipophosphoglycan Increases Macrophage-Dependent Chemotaxis of CXCR6-Expressing Cells via CXCL16 Induction.Infect Immun. 2019 Apr 23;87(5):e00064-19. doi: 10.1128/IAI.00064-19. Print 2019 Mar. Infect Immun. 2019. PMID: 30804103 Free PMC article.
-
Translational Control of Canonical and Non-Canonical Translation Initiation Factors at the Sea Urchin Egg to Embryo Transition.Int J Mol Sci. 2019 Feb 1;20(3):626. doi: 10.3390/ijms20030626. Int J Mol Sci. 2019. PMID: 30717141 Free PMC article.
-
Exploring the potential of dietary factors and plant extracts as chemopreventive agents in oral squamous cell carcinoma treatment.Front Oral Health. 2023 Oct 4;4:1246873. doi: 10.3389/froh.2023.1246873. eCollection 2023. Front Oral Health. 2023. PMID: 37859687 Free PMC article. Review.
-
Non-genomic mechanisms mediate androgen-induced PSD95 expression.Aging (Albany NY). 2019 Apr 20;11(8):2281-2294. doi: 10.18632/aging.101913. Aging (Albany NY). 2019. PMID: 31005955 Free PMC article.
References
-
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG II, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. 2011. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 7:e1001393. doi:10.1371/journal.pgen.1001393. - DOI - PMC - PubMed
-
- Mathews MB, Sonenberg N, Hershey JWB (ed). 2007. Translational control in biology and medicine, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
