Investigating Colonization of the Healthy Adult Gastrointestinal Tract by Fungi
- PMID: 29600282
- PMCID: PMC5874442
- DOI: 10.1128/mSphere.00092-18
Investigating Colonization of the Healthy Adult Gastrointestinal Tract by Fungi
Abstract
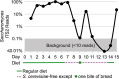
A wide diversity of fungi have been detected in the human gastrointestinal (GI) tract with the potential to provide or influence important functions. However, many of the fungi most commonly detected in stool samples are also present in food or the oral cavity. Therefore, to recognize which gut fungi are likely to have a sustained influence on human health, there is a need to separate transient members of the GI tract from true colonizers. To identify colonizing fungi, the eukaryotic rRNA operon's second internal transcribed spacer (ITS2) was sequenced from the stool, saliva, and food of healthy adults following consumption of different controlled diets. Unlike most bacterial 16S rRNA genes, the only fungal ITS2 operational taxonomic units (OTUs) detected in stool DNA across multiple diets were also present in saliva and/or food. Additional analyses, including culture-based approaches and sequencing of the 18S rRNA gene, ITS2 cDNA, and DNA extracted using alternative methods, failed to detect additional fungi. Two abundant fungi, Saccharomyces cerevisiae and Candida albicans, were examined further in healthy volunteers. Saccharomyces became undetectable in stool when a S. cerevisiae-free diet was consumed, and the levels of C. albicans in stool were dramatically reduced by more frequent cleaning of teeth. Extremely low fungal abundance, the inability of fungi to grow under conditions mimicking the distal gut, and evidence from analysis of other public datasets further support the hypothesis that fungi do not routinely colonize the GI tracts of healthy adults. IMPORTANCE We sought to identify the fungi that colonize healthy GI tracts and that have a sustained influence on the diverse functions of the gut microbiome. Instead, we found that all fungi in the stool of healthy volunteers could be explained by their presence in oral and dietary sources and that our results, together with those from other analyses, support the model that there is little or no gastrointestinal colonization by fungi. This may be due to Westernization, primate evolution, fungal ecology, and/or the strong defenses of a healthy immune system. Importantly, fungal colonization of the GI tract may often be indicative of disease. As fungi can cause serious infections in immunocompromised individuals and are found at increased abundance in multiple disorders of the GI tract, understanding normal fungal colonization is essential for proper treatment and prevention of fungal pathogenesis.
Keywords: colonization; diet; fungi; gastrointestinal tract; mycobiome; saliva.
Figures
Similar articles
-
The gut mycobiome of the Human Microbiome Project healthy cohort.Microbiome. 2017 Nov 25;5(1):153. doi: 10.1186/s40168-017-0373-4. Microbiome. 2017. PMID: 29178920 Free PMC article.
-
Early gut mycobiota and mother-offspring transfer.Microbiome. 2017 Aug 24;5(1):107. doi: 10.1186/s40168-017-0319-x. Microbiome. 2017. PMID: 28837002 Free PMC article. Clinical Trial.
-
Sequence-based methods for detecting and evaluating the human gut mycobiome.Lett Appl Microbiol. 2016 Mar;62(3):209-15. doi: 10.1111/lam.12539. Epub 2016 Feb 2. Lett Appl Microbiol. 2016. PMID: 26669281
-
Fungi in the healthy human gastrointestinal tract.Virulence. 2017 Apr 3;8(3):352-358. doi: 10.1080/21505594.2016.1247140. Epub 2016 Oct 13. Virulence. 2017. PMID: 27736307 Free PMC article. Review.
-
Fungi in Gastrointestinal Tracts of Human and Mice: from Community to Functions.Microb Ecol. 2018 May;75(4):821-829. doi: 10.1007/s00248-017-1105-9. Epub 2017 Nov 6. Microb Ecol. 2018. PMID: 29110065 Review.
Cited by
-
Candida species-specific colonization in the healthy and impaired human gastrointestinal tract as simulated using the Mucosal Ileum-SHIME® model.FEMS Microbiol Ecol. 2024 Aug 13;100(9):fiae113. doi: 10.1093/femsec/fiae113. FEMS Microbiol Ecol. 2024. PMID: 39169462 Free PMC article.
-
Critically Appraising the Significance of the Oral Mycobiome.J Dent Res. 2021 Feb;100(2):133-140. doi: 10.1177/0022034520956975. Epub 2020 Sep 13. J Dent Res. 2021. PMID: 32924741 Free PMC article. Review.
-
Association between ustekinumab therapy and changes in specific anti-microbial response, serum biomarkers, and microbiota composition in patients with IBD: A pilot study.PLoS One. 2022 Dec 30;17(12):e0277576. doi: 10.1371/journal.pone.0277576. eCollection 2022. PLoS One. 2022. PMID: 36584073 Free PMC article.
-
Fecal Microbiome Changes and Specific Anti-Bacterial Response in Patients with IBD during Anti-TNF Therapy.Cells. 2021 Nov 16;10(11):3188. doi: 10.3390/cells10113188. Cells. 2021. PMID: 34831411 Free PMC article.
-
The Role of Lung and Gut Microbiota in the Pathology of Asthma.Immunity. 2020 Feb 18;52(2):241-255. doi: 10.1016/j.immuni.2020.01.007. Immunity. 2020. PMID: 32075727 Free PMC article. Review.
References
-
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336:1314–1317. doi:10.1126/science.1221789. - DOI - PMC - PubMed
-
- Palma ML, Zamith-Miranda D, Martins FS, Bozza FA, Nimrichter L, Montero-Lomeli M, Marques ET Jr, Douradinha B. 2015. Probiotic Saccharomyces cerevisiae strains as biotherapeutic tools: is there room for improvement? Appl Microbiol Biotechnol 99:6563–6570. doi:10.1007/s00253-015-6776-x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
