RING tetramerization is required for nuclear body biogenesis and PML sumoylation
- PMID: 29599493
- PMCID: PMC5876331
- DOI: 10.1038/s41467-018-03498-0
RING tetramerization is required for nuclear body biogenesis and PML sumoylation
Erratum in
-
Publisher Correction: RING tetramerization is required for nuclear body biogenesis and PML sumoylation.Nat Commun. 2018 May 4;9(1):1841. doi: 10.1038/s41467-018-04347-w. Nat Commun. 2018. PMID: 29728567 Free PMC article.
Abstract
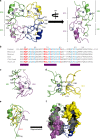
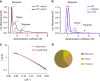
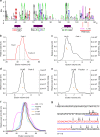
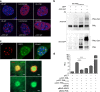
ProMyelocyticLeukemia nuclear bodies (PML NBs) are stress-regulated domains directly implicated in acute promyelocytic leukemia eradication. Most TRIM family members bind ubiquitin E2s and many acquire ligase activity upon RING dimerization. In contrast, PML binds UBC9, the SUMO E2 enzyme. Here, using X-ray crystallography and SAXS characterization, we demonstrate that PML RING tetramerizes through highly conserved PML-specific sequences, which are required for NB assembly and PML sumoylation. Conserved residues implicated in RING dimerization of other TRIMs also contribute to PML tetramer stability. Wild-type PML rescues the ability of some RING mutants to form NBs as well as their sumoylation. Impaired RING tetramerization abolishes PML/RARA-driven leukemogenesis in vivo and arsenic-induced differentiation ex vivo. Our studies thus identify RING tetramerization as a key step in the NB macro-molecular scaffolding. They suggest that higher order RING interactions allow efficient UBC9 recruitment and thus change the biochemical nature of TRIM-facilitated post-translational modifications.
Conflict of interest statement
All authors declare no competing interests.
Figures

Similar articles
-
Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins.J Cell Biol. 2014 Mar 17;204(6):931-45. doi: 10.1083/jcb.201305148. J Cell Biol. 2014. PMID: 24637324 Free PMC article.
-
Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9.Sci Rep. 2018 May 17;8(1):7754. doi: 10.1038/s41598-018-25150-z. Sci Rep. 2018. PMID: 29773808 Free PMC article.
-
SUMOylation regulates the number and size of promyelocytic leukemia-nuclear bodies (PML-NBs) and arsenic perturbs SUMO dynamics on PML by insolubilizing PML in THP-1 cells.Arch Toxicol. 2022 Feb;96(2):545-558. doi: 10.1007/s00204-021-03195-w. Epub 2022 Jan 10. Arch Toxicol. 2022. PMID: 35001170
-
Unravelling the molecular interplay: SUMOylation, PML nuclear bodies and vascular cell activity in health and disease.Cell Signal. 2024 Jul;119:111156. doi: 10.1016/j.cellsig.2024.111156. Epub 2024 Apr 2. Cell Signal. 2024. PMID: 38574938 Review.
-
A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics.Int J Biol Sci. 2010 Jan 12;6(1):51-67. doi: 10.7150/ijbs.6.51. Int J Biol Sci. 2010. PMID: 20087442 Free PMC article. Review.
Cited by
-
Interplay between RNA Viruses and Promyelocytic Leukemia Nuclear Bodies.Vet Sci. 2021 Mar 31;8(4):57. doi: 10.3390/vetsci8040057. Vet Sci. 2021. PMID: 33807177 Free PMC article. Review.
-
SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes.Mol Cell. 2018 Aug 2;71(3):409-418. doi: 10.1016/j.molcel.2018.07.027. Mol Cell. 2018. PMID: 30075142 Free PMC article. Review.
-
The B-box1 domain of PML mediates SUMO E2-E3 complex formation through an atypical interaction with UBC9.Biophys Chem. 2022 Aug;287:106827. doi: 10.1016/j.bpc.2022.106827. Epub 2022 May 18. Biophys Chem. 2022. PMID: 35667129 Free PMC article.
-
Multiple roles of arsenic compounds in phase separation and membraneless organelles formation determine their therapeutic efficacy in tumors.J Pharm Anal. 2024 Aug;14(8):100957. doi: 10.1016/j.jpha.2024.02.011. Epub 2024 Feb 24. J Pharm Anal. 2024. PMID: 39253293 Free PMC article. Review.
-
PML Nuclear bodies: the cancer connection and beyond.Nucleus. 2024 Dec;15(1):2321265. doi: 10.1080/19491034.2024.2321265. Epub 2024 Feb 27. Nucleus. 2024. PMID: 38411156 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

