Deaminase-Dead Mouse APOBEC3 Is an In Vivo Retroviral Restriction Factor
- PMID: 29593034
- PMCID: PMC5952145
- DOI: 10.1128/JVI.00168-18
Deaminase-Dead Mouse APOBEC3 Is an In Vivo Retroviral Restriction Factor
Abstract
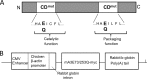
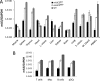
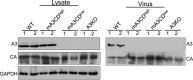
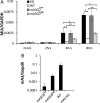
The apolipoprotein B editing complex 3 (APOBEC3) proteins are potent retroviral restriction factors that are under strong positive selection, both in terms of gene copy number and sequence diversity. A common feature of all the members of the APOBEC3 family is the presence of one or two cytidine deamination domains, essential for cytidine deamination of retroviral reverse transcripts as well as packaging into virions. Several studies have indicated that human and mouse APOBEC3 proteins restrict retrovirus infection via cytidine deaminase (CD)-dependent and -independent means. To understand the relative contribution of CD-independent restriction in vivo, we created strains of transgenic mice on an APOBEC3 knockout background that express a deaminase-dead mouse APOBEC3 due to point mutations in both CD domains (E73Q/E253Q). Here, we show that the CD-dead APOBEC3 can restrict murine retroviruses in vivo Moreover, unlike the wild-type protein, the mutant APOBEC3 is not packaged into virions but acts only as a cell-intrinsic restriction factor that blocks reverse transcription by incoming viruses. Finally, we show that wild-type and CD-dead mouse APOBEC3 can bind to murine leukemia virus (MLV) reverse transcriptase. Our findings suggest that the mouse APOBEC3 cytidine deaminase activity is not required for retrovirus restriction.IMPORTANCE APOBEC3 proteins are important host cellular restriction factors essential for restricting retrovirus infection by causing mutations in the virus genome and by blocking reverse transcription. While both methods of restriction function in vitro, little is known about their role during in vivo infection. By developing transgenic mice with mutations in the cytidine deamination domains needed for enzymatic activity and interaction with viral RNA, we show that APOBEC3 proteins can still restrict in vivo infection by interacting with reverse transcriptase and blocking its activity. These studies demonstrate that APOBEC3 proteins have evolved multiple means for blocking retrovirus infection and that all of these means function in vivo.
Keywords: APOBEC3; MLV; cytidine deamination domains; retrovirus; transgenic mice; virus pathogenesis.
Copyright © 2018 American Society for Microbiology.
Figures


Similar articles
-
Mouse APOBEC3 Restriction of Retroviruses.Viruses. 2020 Oct 27;12(11):1217. doi: 10.3390/v12111217. Viruses. 2020. PMID: 33121095 Free PMC article. Review.
-
Murine Leukemia Virus P50 Protein Counteracts APOBEC3 by Blocking Its Packaging.J Virol. 2020 Aug 31;94(18):e00032-20. doi: 10.1128/JVI.00032-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641479 Free PMC article.
-
APOBEC3 inhibition of mouse mammary tumor virus infection: the role of cytidine deamination versus inhibition of reverse transcription.J Virol. 2013 May;87(9):4808-17. doi: 10.1128/JVI.00112-13. Epub 2013 Feb 28. J Virol. 2013. PMID: 23449789 Free PMC article.
-
Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex.Proc Natl Acad Sci U S A. 2013 May 28;110(22):9078-83. doi: 10.1073/pnas.1217399110. Epub 2013 May 13. Proc Natl Acad Sci U S A. 2013. PMID: 23671100 Free PMC article.
-
Hepatitis B: modern concepts in pathogenesis--APOBEC3 cytidine deaminases as effectors in innate immunity against the hepatitis B virus.Curr Opin Infect Dis. 2008 Jun;21(3):298-303. doi: 10.1097/QCO.0b013e3282fe1bb2. Curr Opin Infect Dis. 2008. PMID: 18448976 Review.
Cited by
-
Mouse APOBEC3 Restriction of Retroviruses.Viruses. 2020 Oct 27;12(11):1217. doi: 10.3390/v12111217. Viruses. 2020. PMID: 33121095 Free PMC article. Review.
-
Potential Role of APOBEC3 Family Proteins in SARS-CoV-2 Replication.Viruses. 2024 Jul 16;16(7):1141. doi: 10.3390/v16071141. Viruses. 2024. PMID: 39066304 Free PMC article.
-
SERINC5 Potently Restricts Retrovirus Infection In Vivo.mBio. 2020 Jul 14;11(4):e00588-20. doi: 10.1128/mBio.00588-20. mBio. 2020. PMID: 32665269 Free PMC article.
-
Murine Leukemia Virus P50 Protein Counteracts APOBEC3 by Blocking Its Packaging.J Virol. 2020 Aug 31;94(18):e00032-20. doi: 10.1128/JVI.00032-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641479 Free PMC article.
-
Elucidating the Antiviral Mechanism of Different MARCH Factors.mBio. 2021 Mar 2;12(2):e03264-20. doi: 10.1128/mBio.03264-20. mBio. 2021. PMID: 33653895 Free PMC article.
References
-
- Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. 2010. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog 6:e1000974. doi:10.1371/journal.ppat.1000974. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

