Posttranslational modifications of CENP-A: marks of distinction
- PMID: 29569072
- PMCID: PMC6082721
- DOI: 10.1007/s00412-018-0665-x
Posttranslational modifications of CENP-A: marks of distinction
Abstract
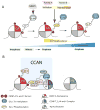
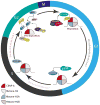
Centromeres are specialized chromosome domain that serve as the site for kinetochore assembly and microtubule attachment during cell division, to ensure proper segregation of chromosomes. In higher eukaryotes, the identity of active centromeres is marked by the presence of CENP-A (centromeric protein-A), a histone H3 variant. CENP-A forms a centromere-specific nucleosome that acts as a foundation for centromere assembly and function. The posttranslational modification (PTM) of histone proteins is a major mechanism regulating the function of chromatin. While a few CENP-A site-specific modifications are shared with histone H3, the majority are specific to CENP-A-containing nucleosomes, indicating that modification of these residues contribute to centromere-specific function. CENP-A undergoes posttranslational modifications including phosphorylation, acetylation, methylation, and ubiquitylation. Work from many laboratories have uncovered the importance of these CENP-A modifications in its deposition at centromeres, protein stability, and recruitment of the CCAN (constitutive centromere-associated network). Here, we discuss the PTMs of CENP-A and their biological relevance.
Keywords: CENP-A; Centromere; Chromatin; Kinetochore; Mitosis; Posttranslational modification.
Figures



Similar articles
-
Transcription of a centromere-enriched retroelement and local retention of its RNA are significant features of the CENP-A chromatin landscape.Genome Biol. 2024 Nov 18;25(1):295. doi: 10.1186/s13059-024-03433-1. Genome Biol. 2024. PMID: 39558354 Free PMC article.
-
Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP.Cell. 2009 May 1;137(3):472-84. doi: 10.1016/j.cell.2009.02.039. Cell. 2009. PMID: 19410544 Free PMC article.
-
Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini.Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16588-93. doi: 10.1073/pnas.1113621108. Epub 2011 Sep 26. Proc Natl Acad Sci U S A. 2011. PMID: 21949362 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
The effectiveness of abstinence-based and harm reduction-based interventions in reducing problematic substance use in adults who are experiencing homelessness in high income countries: A systematic review and meta-analysis: A systematic review.Campbell Syst Rev. 2024 Apr 21;20(2):e1396. doi: 10.1002/cl2.1396. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38645303 Free PMC article. Review.
Cited by
-
Histone N-alpha terminal modifications: genome regulation at the tip of the tail.Epigenetics Chromatin. 2020 Jul 17;13(1):29. doi: 10.1186/s13072-020-00352-w. Epigenetics Chromatin. 2020. PMID: 32680559 Free PMC article. Review.
-
The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle.Open Biol. 2020 Jun;10(6):200051. doi: 10.1098/rsob.200051. Epub 2020 Jun 10. Open Biol. 2020. PMID: 32516549 Free PMC article. Review.
-
Guarding the Genome: CENP-A-Chromatin in Health and Cancer.Genes (Basel). 2020 Jul 16;11(7):810. doi: 10.3390/genes11070810. Genes (Basel). 2020. PMID: 32708729 Free PMC article. Review.
-
Kinetochore assembly throughout the cell cycle.Semin Cell Dev Biol. 2021 Sep;117:62-74. doi: 10.1016/j.semcdb.2021.03.008. Epub 2021 Mar 19. Semin Cell Dev Biol. 2021. PMID: 33753005 Free PMC article.
-
Pan-Cancer and Single-Cell Analysis Reveals CENPL as a Cancer Prognosis and Immune Infiltration-Related Biomarker.Front Immunol. 2022 Jun 30;13:916594. doi: 10.3389/fimmu.2022.916594. eCollection 2022. Front Immunol. 2022. PMID: 35844598 Free PMC article.
References
-
- Bade D, Pauleau AL, Wendler A, Erhardt S. The E3 ligase CUL3/ RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell. 2014;28:508–519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

