Cis- and Trans-Modifiers of Repeat Expansions: Blending Model Systems with Human Genetics
- PMID: 29567336
- PMCID: PMC5959756
- DOI: 10.1016/j.tig.2018.02.005
Cis- and Trans-Modifiers of Repeat Expansions: Blending Model Systems with Human Genetics
Abstract
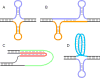
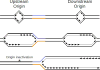
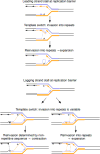
Over 30 hereditary diseases are caused by the expansion of microsatellite repeats. The length of the expandable repeat is the main hereditary determinant of these disorders. They are also affected by numerous genomic variants that are either nearby (cis) or physically separated from (trans) the repetitive locus, which we review here. These genetic variants have largely been elucidated in model systems using gene knockouts, while a few have been directly observed as single-nucleotide polymorphisms (SNPs) in patients. There is a notable disconnect between these two bodies of knowledge: knockouts poorly approximate the SNP-level variation in human populations that gives rise to medically relevant cis- and trans-modifiers, while the rarity of these diseases limits the statistical power of SNP-based analysis in humans. We propose that high-throughput SNP-based screening in model systems could become a useful approach to quickly identify and characterize modifiers of clinical relevance for patients.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
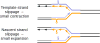

Similar articles
-
Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications.Nat Rev Genet. 2024 Jul;25(7):476-499. doi: 10.1038/s41576-024-00696-z. Epub 2024 Mar 11. Nat Rev Genet. 2024. PMID: 38467784 Review.
-
On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability.J Biol Chem. 2020 Mar 27;295(13):4134-4170. doi: 10.1074/jbc.REV119.007678. Epub 2020 Feb 14. J Biol Chem. 2020. PMID: 32060097 Free PMC article. Review.
-
Triplet repeat length bias and variation in the human transcriptome.Proc Natl Acad Sci U S A. 2009 Oct 6;106(40):17095-100. doi: 10.1073/pnas.0907112106. Epub 2009 Sep 17. Proc Natl Acad Sci U S A. 2009. PMID: 19805156 Free PMC article.
-
Genome-wide detection of tandem DNA repeats that are expanded in autism.Nature. 2020 Oct;586(7827):80-86. doi: 10.1038/s41586-020-2579-z. Epub 2020 Jul 27. Nature. 2020. PMID: 32717741 Free PMC article.
-
STRetch: detecting and discovering pathogenic short tandem repeat expansions.Genome Biol. 2018 Aug 21;19(1):121. doi: 10.1186/s13059-018-1505-2. Genome Biol. 2018. PMID: 30129428 Free PMC article.
Cited by
-
Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications.Nat Rev Genet. 2024 Jul;25(7):476-499. doi: 10.1038/s41576-024-00696-z. Epub 2024 Mar 11. Nat Rev Genet. 2024. PMID: 38467784 Review.
-
Expanding horizons of tandem repeats in biology and medicine: Why 'genomic dark matter' matters.Emerg Top Life Sci. 2023 Dec 13;7(3):239-47. doi: 10.1042/ETLS20230075. Online ahead of print. Emerg Top Life Sci. 2023. PMID: 38088823 Free PMC article.
-
Elevated MSH2 MSH3 expression interferes with DNA metabolism in vivo.Nucleic Acids Res. 2023 Dec 11;51(22):12185-12206. doi: 10.1093/nar/gkad934. Nucleic Acids Res. 2023. PMID: 37930834 Free PMC article.
-
Challenges facing repeat expansion identification, characterisation, and the pathway to discovery.Emerg Top Life Sci. 2023 Dec 14;7(3):339-348. doi: 10.1042/ETLS20230019. Emerg Top Life Sci. 2023. PMID: 37888797 Free PMC article. Review.
-
Mutation and selection processes regulating short tandem repeats give rise to genetic and phenotypic diversity across species.J Evol Biol. 2023 Feb;36(2):321-336. doi: 10.1111/jeb.14106. Epub 2022 Oct 26. J Evol Biol. 2023. PMID: 36289560 Free PMC article. Review.
References
-
- Pearson CE, et al. Repeat instability: Mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. - PubMed
-
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. - PubMed
-
- Orr HT, Zoghbi HY. Trinucleotide Repeat Disorders. Annu. Rev. Neurosci. 2007;30:575–621. - PubMed
-
- Monckton DG, et al. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet. 1995;4:1–8. - PubMed
-
- De Biase I, et al. Progressive GAA expansions in dorsal root ganglia of Friedreich’s ataxia patients. Ann. Neurol. 2007;61:55–60. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

