Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree
- PMID: 29563566
- PMCID: PMC5862945
- DOI: 10.1038/s41598-018-23094-y
Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree
Abstract
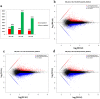
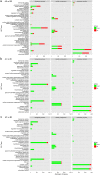
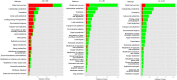
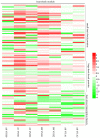
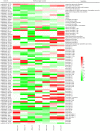
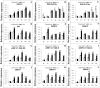
Two contrasting cold response rubber tree clones, the cold-resistant '93-114' and cold-sensitive 'Reken501', were subject to a global transcriptome response assessing via high-throughput RNA-seq technique and comprehensive bioinformatics analysis using the referenced rubber tree genome with the purpose of exploring the potential molecular cues underlying the tolerance of rubber trees to cold stress. As a result, a total of 1919 genes had significantly higher expression, while 2929 genes had significantly lower expression in '93-114' than in 'Reken501' without cold stress. Upon cold stress, the numbers of genes with significantly higher expression decreased to 1501 at 1 h treatment and to 1285 at 24 h treatment in '93-114' than that of 'Reken501', conversely, the numbers of genes with significantly lower expression increased to 7567 at 1 h treatment and to 5482 at 24 h treatment. Functional annotation of the differentially expressed genes between '93-114' and 'Reken501' suggests that down-regulation of auxin and ethylene signaling and activation of heat shock module and ROS scavengers is a primary strategy for H. brasiliensis to cope with cold stress. Our identified vital differentially expressed genes may be beneficial for elucidation of the molecular mechanisms underlying cold tolerance and for genetic improvement of H. brasiliensis clones.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Comparative transcriptome analysis reveals an early gene expression profile that contributes to cold resistance in Hevea brasiliensis (the Para rubber tree).Tree Physiol. 2018 Sep 1;38(9):1409-1423. doi: 10.1093/treephys/tpy014. Tree Physiol. 2018. PMID: 29474681
-
Deep expression analysis reveals distinct cold-response strategies in rubber tree (Hevea brasiliensis).BMC Genomics. 2019 Jun 4;20(1):455. doi: 10.1186/s12864-019-5852-5. BMC Genomics. 2019. PMID: 31164105 Free PMC article.
-
Transcriptome profiling of rubber tree (Hevea brasiliensis) discovers candidate regulators of the cold stress response.Genes Genomics. 2018 Nov;40(11):1181-1197. doi: 10.1007/s13258-018-0681-5. Epub 2018 Mar 6. Genes Genomics. 2018. PMID: 30315521
-
Cold signal transduction and its interplay with phytohormones during cold acclimation.Plant Cell Physiol. 2015 Jan;56(1):7-15. doi: 10.1093/pcp/pcu115. Epub 2014 Sep 3. Plant Cell Physiol. 2015. PMID: 25189343 Review.
-
Heat stress in Marchantia polymorpha: Sensing and mechanisms underlying a dynamic response.Plant Cell Environ. 2021 Jul;44(7):2134-2149. doi: 10.1111/pce.13914. Epub 2020 Oct 26. Plant Cell Environ. 2021. PMID: 33058168 Review.
Cited by
-
Role of jasmonic acid in plants: the molecular point of view.Plant Cell Rep. 2021 Aug;40(8):1471-1494. doi: 10.1007/s00299-021-02687-4. Epub 2021 Apr 5. Plant Cell Rep. 2021. PMID: 33821356 Review.
-
Transcriptomics integrated with widely targeted metabolomics reveals the cold resistance mechanism in Hevea brasiliensis.Front Plant Sci. 2023 Jan 10;13:1092411. doi: 10.3389/fpls.2022.1092411. eCollection 2022. Front Plant Sci. 2023. PMID: 36704172 Free PMC article.
-
The rubber tree kinome: Genome-wide characterization and insights into coexpression patterns associated with abiotic stress responses.Front Plant Sci. 2023 Feb 7;14:1068202. doi: 10.3389/fpls.2023.1068202. eCollection 2023. Front Plant Sci. 2023. PMID: 36824205 Free PMC article.
-
Gene-coexpression network analysis identifies specific modules and hub genes related to cold stress in rice.BMC Genomics. 2022 Apr 1;23(1):251. doi: 10.1186/s12864-022-08438-3. BMC Genomics. 2022. PMID: 35365095 Free PMC article.
-
Genomic Characteristics and Comparative Genomics Analysis of Two Chinese Corynespora cassiicola Strains Causing Corynespora Leaf Fall (CLF) Disease.J Fungi (Basel). 2021 Jun 16;7(6):485. doi: 10.3390/jof7060485. J Fungi (Basel). 2021. PMID: 34208763 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

