First Evidence for Internal Ribosomal Entry Sites in Diverse Fungal Virus Genomes
- PMID: 29559577
- PMCID: PMC5874917
- DOI: 10.1128/mBio.02350-17
First Evidence for Internal Ribosomal Entry Sites in Diverse Fungal Virus Genomes
Abstract
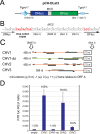
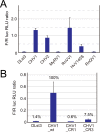
In contrast to well-established internal ribosomal entry site (IRES)-mediated translational initiation in animals and plants, no IRESs were established in fungal viral or cellular RNAs. To identify IRES elements in mycoviruses, we developed a luciferase-based dual-reporter detection system in Cryphonectria parasitica, a model filamentous fungus for virus-host interactions. A bicistronic construct entails a codon-optimized Renilla and firefly luciferase (ORluc and OFluc, respectively) gene, between which potential IRES sequences can be inserted. In this system, ORluc serves as an internal control, while OFluc represents IRES activity. Virus sequences in the 5' untranslated regions (UTRs) of the genomes of diverse positive-sense single-stranded RNA and double-stranded RNA (dsRNA) viruses were analyzed. The results show relatively high IRES activities for Cryphonectria hypovirus 1 (CHV1) and CHV2 and faint but measurable activity for CHV3. The weak IRES signal of CHV3 may be explained by its monocistronic nature, differing from the bicistronic nature of CHV1 and CHV2. This would allow these three hypoviruses to have similar rates of translation of replication-associated protein per viral mRNA molecule. The importance of 24 5'-proximal codons of CHV1 as well as the 5' UTR for IRES function was confirmed. Furthermore, victoriviruses and chrysoviruses tested IRES positive, whereas mycoreoviruses, partitiviruses, and quadriviruses showed similar Fluc activities as the negative controls. Overall, this study represents the first development of an IRES identification system in filamentous fungi based on the codon-optimized dual-luciferase assay and provides evidence for IRESs in filamentous fungi.IMPORTANCE Cap-independent, internal ribosomal entry site (IRES)-mediated translational initiation is often used by virus mRNAs and infrequently by cellular mRNAs in animals and plants. However, no IRESs have been established in fungal virus RNAs or cellular RNAs in filamentous fungi. Here, we report the development of a dual-luciferase assay system and measurement of the IRES activities of fungal RNA viruses in a model filamentous fungal host, Cryphonectria parasitica Viruses identified as IRES positive include hypoviruses (positive-sense RNA viruses, members of the expanded Picornavirus supergroup), totiviruses (nonsegmented dsRNA viruses), and chrysoviruses (tetrasegmented dsRNA viruses). No IRES activities were observed in the 5' untranslated regions of mycoreoviruses (11-segmented dsRNA viruses), quadriviruses (tetrasegmented dsRNA viruses), or partitiviruses (bisegmented dsRNA viruses). This study provides the first evidence for IRES activities in diverse RNA viruses in filamentous fungi and is a first step toward identifying trans-acting host factors and cis-regulatory viral RNA elements.
Keywords: dsRNA virus; hypovirus; internal ribosome entry site; mycovirus; noncanonical translation.
Copyright © 2018 Chiba et al.
Figures

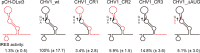

Similar articles
-
Effect of intercistronic length on internal ribosome entry site (IRES) efficiency in bicistronic mRNA.Gene Expr. 1999;8(5-6):299-309. Gene Expr. 1999. PMID: 10947079 Free PMC article.
-
A Sequence-Independent, Unstructured Internal Ribosome Entry Site Is Responsible for Internal Expression of the Coat Protein of Turnip Crinkle Virus.J Virol. 2017 Mar 29;91(8):e02421-16. doi: 10.1128/JVI.02421-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28179526 Free PMC article.
-
IRES-mediated translation of foot-and-mouth disease virus (FMDV) in cultured cells derived from FMDV-susceptible and -insusceptible animals.BMC Vet Res. 2016 Mar 31;12:66. doi: 10.1186/s12917-016-0694-8. BMC Vet Res. 2016. PMID: 27036295 Free PMC article.
-
Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review.
-
IRES-mediated cap-independent translation, a path leading to hidden proteome.J Mol Cell Biol. 2019 Oct 25;11(10):911-919. doi: 10.1093/jmcb/mjz091. J Mol Cell Biol. 2019. PMID: 31504667 Free PMC article. Review.
Cited by
-
A Transfectable Fusagravirus from a Japanese Strain of Cryphonectria carpinicola with Spherical Particles.Viruses. 2022 Aug 4;14(8):1722. doi: 10.3390/v14081722. Viruses. 2022. PMID: 36016344 Free PMC article.
-
CpSmt3, an ortholog of small ubiquitin-like modifier, is essential for growth, organelle function, virulence, and antiviral defense in Cryphonectria parasitica.Front Microbiol. 2024 May 9;15:1391855. doi: 10.3389/fmicb.2024.1391855. eCollection 2024. Front Microbiol. 2024. PMID: 38784801 Free PMC article.
-
Dicer functions transcriptionally and posttranscriptionally in a multilayer antiviral defense.Proc Natl Acad Sci U S A. 2019 Feb 5;116(6):2274-2281. doi: 10.1073/pnas.1812407116. Epub 2019 Jan 23. Proc Natl Acad Sci U S A. 2019. PMID: 30674672 Free PMC article.
-
Identification of a Novel Hypovirulence-Inducing Hypovirus From Alternaria alternata.Front Microbiol. 2019 May 15;10:1076. doi: 10.3389/fmicb.2019.01076. eCollection 2019. Front Microbiol. 2019. PMID: 31156589 Free PMC article.
-
Xrn1-resistant RNA motifs are disseminated throughout the RNA virome and are able to block scanning ribosomes.Sci Rep. 2023 Sep 25;13(1):15987. doi: 10.1038/s41598-023-43001-4. Sci Rep. 2023. PMID: 37749116 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
