Divergent transcriptomic responses underlying the ranaviruses-amphibian interaction processes on interspecies infection of Chinese giant salamander
- PMID: 29558886
- PMCID: PMC5861657
- DOI: 10.1186/s12864-018-4596-y
Divergent transcriptomic responses underlying the ranaviruses-amphibian interaction processes on interspecies infection of Chinese giant salamander
Abstract
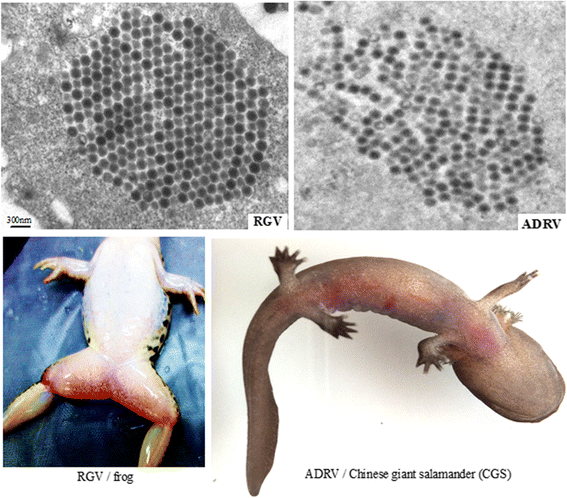
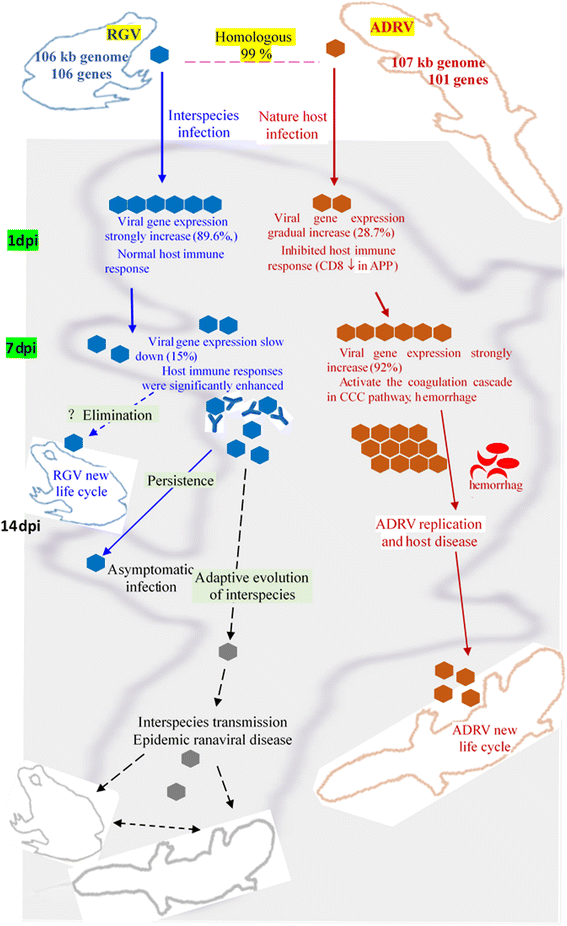
Background: Ranaviruses (family Iridoviridae, nucleocytoplasmic large DNA viruses) have been reported as promiscuous pathogens of cold-blooded vertebrates. Rana grylio virus (RGV, a ranavirus), from diseased frog Rana grylio with a genome of 105.79 kb and Andrias davidianus ranavirus (ADRV), from diseased Chinese giant salamander (CGS) with a genome of 106.73 kb, contains 99% homologous genes.
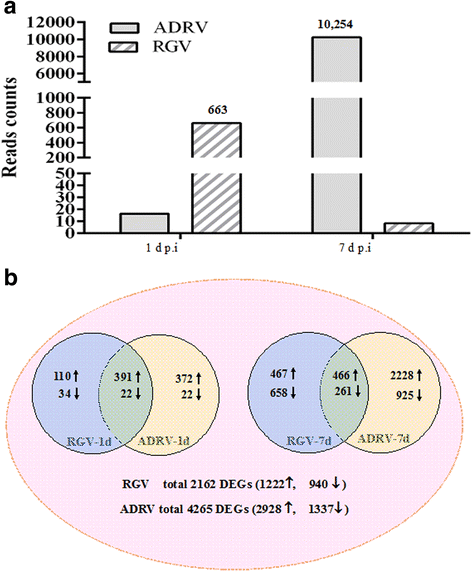
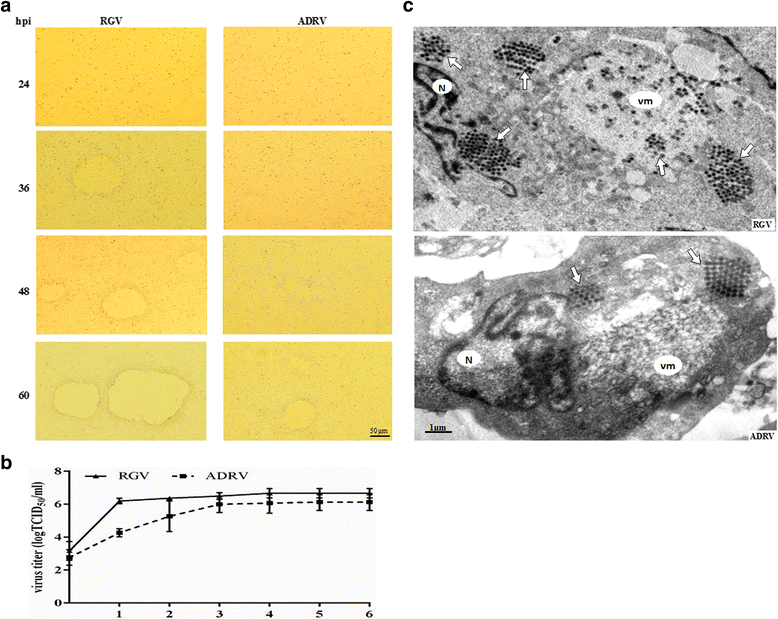
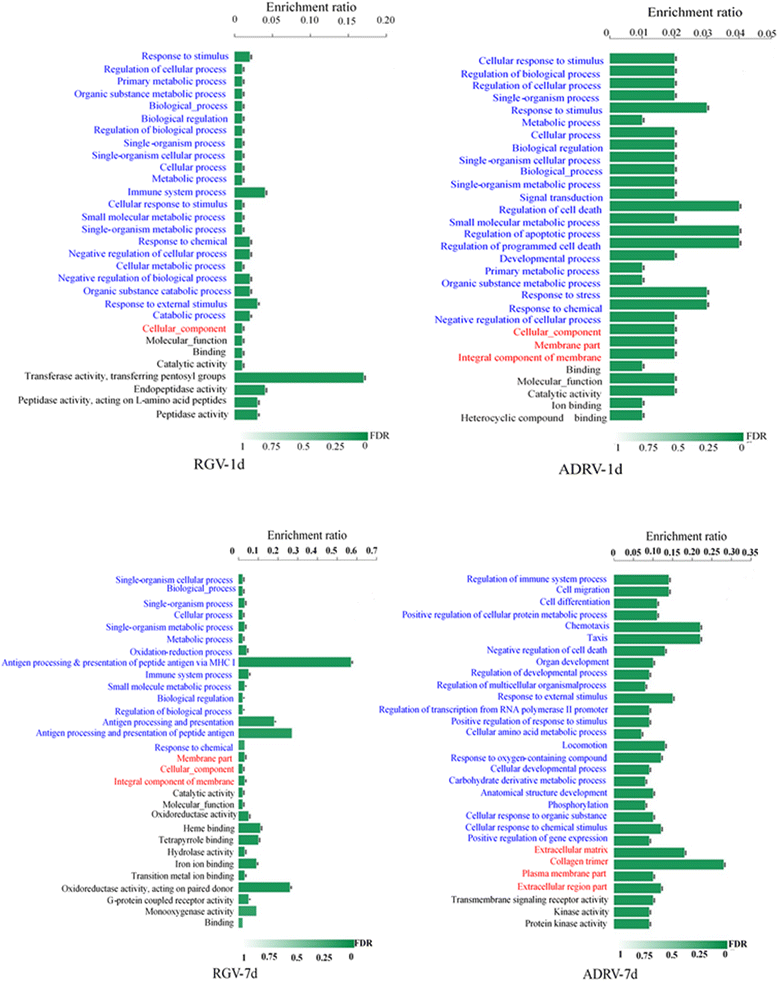
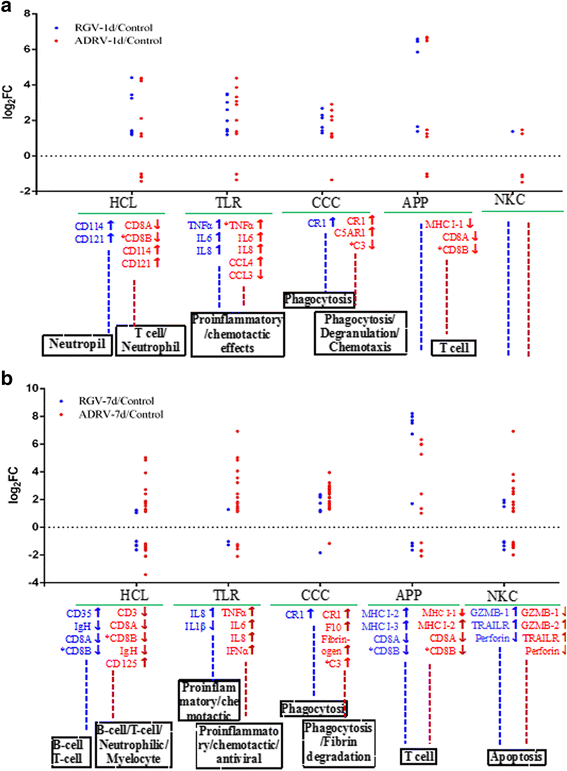
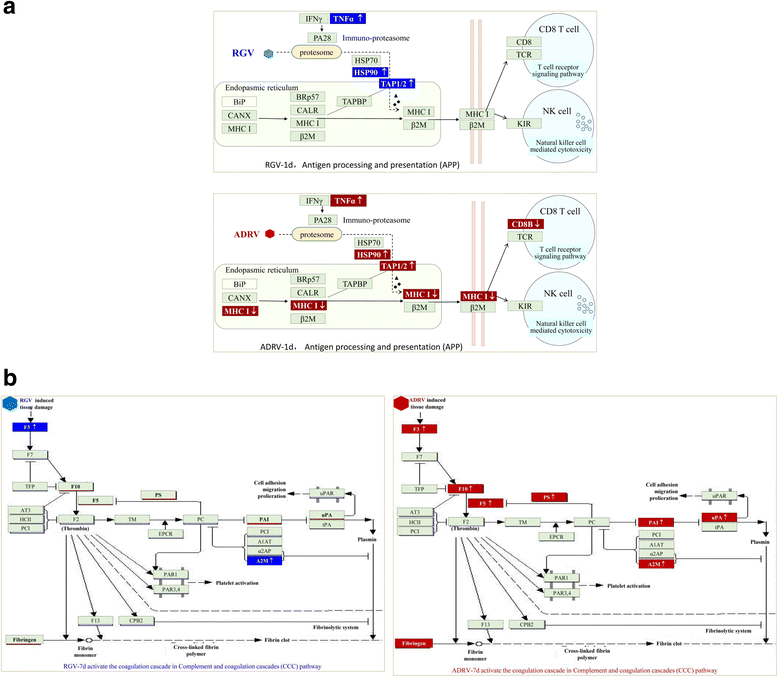
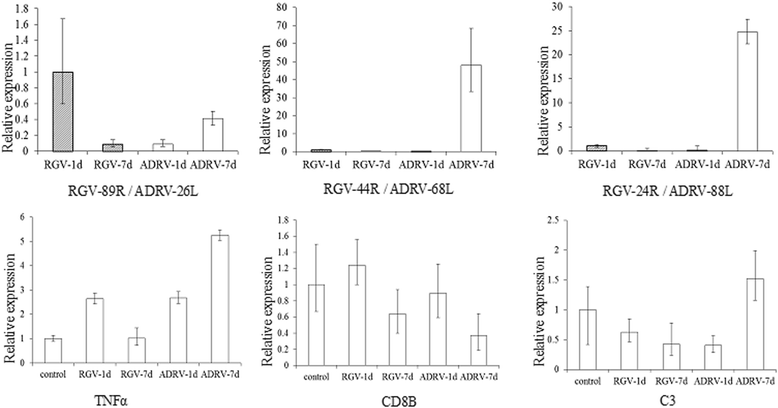
Results: To uncover the differences in virus replication and host responses under interspecies infection, we analyzed transcriptomes of CGS challenged with RGV and ADRV in different time points (1d, 7d) for the first time. A total of 128,533 unigenes were obtained from 820,858,128 clean reads. Transcriptome analysis revealed stronger gene expression of RGV than ADRV at 1 d post infection (dpi), which was supported by infection in vitro. RGV replicated faster and had higher titers than ADRV in cultured CGS cell line. RT-qPCR revealed the RGV genes including the immediate early gene (RGV-89R) had higher expression level than that of ADRV at 1 dpi. It further verified the acute infection of RGV in interspecies infection. The number of differentially expressed genes and enriched pathways from RGV were lower than that from ADRV, which reflected the variant host responses at transcriptional level. No obvious changes of key components in pathway "Antigen processing and presentation" were detected for RGV at 1 dpi. Contrarily, ADRV infection down-regulated the expression levels of MHC I and CD8. The divergent host immune responses revealed the differences between interspecies and natural infection, which may resulted in different fates of the two viruses. Altogether, these results revealed the differences in transcriptome responses among ranavirus interspecies infection of amphibian and new insights in DNA virus-host interactions in interspecies infection.
Conclusion: The DNA virus (RGV) not only expressed self-genes and replicated quickly after entry into host under interspecies infection, but also avoided the over-activation of host responses. The strategy could gain time for the survival of interspecies pathogen, and may provide opportunity for its adaptive evolution and interspecies transmission.
Keywords: Amphibian; Chinese giant salamander; Immune pathway; Interspecies infection; Ranavirus; Transcriptome response; Virus-host interactions.
Conflict of interest statement
Ethics approval
Cultured CGSs were obtained from a farm in Jiangxi, China. The permission for collecting the specimens is not needed in the present study. The specimens are aquaculture animals. The animal procedure and protocol was approved by the Institutional Animal Care and Use Committee of the Institute of Hydrobiology, Chinese Academy of Sciences (Approval number: Y51317-1-301). The statements have been added in the ‘Ethics Approval and Consent to Participate’ section. All surgery was performed under the efforts made to minimize potential harmful effects.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
The Immune System and the Antiviral Responses in Chinese Giant Salamander, Andrias davidianus.Front Immunol. 2021 Oct 5;12:718627. doi: 10.3389/fimmu.2021.718627. eCollection 2021. Front Immunol. 2021. PMID: 34675918 Free PMC article. Review.
-
Genome architecture changes and major gene variations of Andrias davidianus ranavirus (ADRV).Vet Res. 2013 Oct 21;44(1):101. doi: 10.1186/1297-9716-44-101. Vet Res. 2013. PMID: 24143877 Free PMC article.
-
Ranaviruses Bind Cells from Different Species through Interaction with Heparan Sulfate.Viruses. 2019 Jun 29;11(7):593. doi: 10.3390/v11070593. Viruses. 2019. PMID: 31261956 Free PMC article.
-
Protective Immunity Induced by DNA Vaccination against Ranavirus Infection in Chinese Giant Salamander Andrias davidianus.Viruses. 2018 Jan 24;10(2):52. doi: 10.3390/v10020052. Viruses. 2018. PMID: 29364850 Free PMC article.
-
The molecular biology of frog virus 3 and other iridoviruses infecting cold-blooded vertebrates.Viruses. 2011 Oct;3(10):1959-85. doi: 10.3390/v3101959. Epub 2011 Oct 20. Viruses. 2011. PMID: 22069524 Free PMC article. Review.
Cited by
-
Interaction between Two Iridovirus Core Proteins and Their Effects on Ranavirus (RGV) Replication in Cells from Different Species.Viruses. 2019 May 4;11(5):416. doi: 10.3390/v11050416. Viruses. 2019. PMID: 31060251 Free PMC article.
-
Identification of two ISG15 homologues involved in host immune response against RGNNV in Asian seabass (Lates calcarifer).Fish Shellfish Immunol Rep. 2022 Mar 8;3:100054. doi: 10.1016/j.fsirep.2022.100054. eCollection 2022 Dec. Fish Shellfish Immunol Rep. 2022. PMID: 36419602 Free PMC article.
-
The Immune System and the Antiviral Responses in Chinese Giant Salamander, Andrias davidianus.Front Immunol. 2021 Oct 5;12:718627. doi: 10.3389/fimmu.2021.718627. eCollection 2021. Front Immunol. 2021. PMID: 34675918 Free PMC article. Review.
-
Environmental Factors and Their Threshold Affecting the Survival of Five Aquatic Animal Viruses in Different Animal Cells.Viruses. 2022 Nov 17;14(11):2546. doi: 10.3390/v14112546. Viruses. 2022. PMID: 36423155 Free PMC article.
-
Andrias davidianus Ranavirus (ADRV) Genome Replicate Efficiently by Engaging Cellular Mismatch Repair Protein MSH2.Viruses. 2022 May 2;14(5):952. doi: 10.3390/v14050952. Viruses. 2022. PMID: 35632694 Free PMC article.
References
-
- Chan JF, Chan KH, Choi KY, To KKW. Tse H, Cai J, et al. Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg Microbes Infect. 2016;5:e93. doi: 10.1038/emi.2016.99. - DOI - PMC - PubMed
-
- Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza a viruses. Curr Top Microbiol Immunol. 2014;385:359–375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

