Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors
- PMID: 29557770
- PMCID: PMC6537626
- DOI: 10.1099/jgv.0.001047
Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors
Abstract
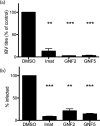
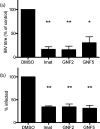
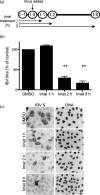
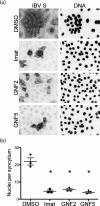
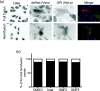
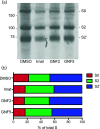
Enveloped viruses gain entry into host cells by fusing with cellular membranes, a step that is required for virus replication. Coronaviruses, including the severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and infectious bronchitis virus (IBV), fuse at the plasma membrane or use receptor-mediated endocytosis and fuse with endosomes, depending on the cell or tissue type. The virus spike (S) protein mediates fusion with the host cell membrane. We have shown previously that an Abelson (Abl) kinase inhibitor, imatinib, significantly reduces SARS-CoV and MERS-CoV viral titres and prevents endosomal entry by HIV SARS S and MERS S pseudotyped virions. SARS-CoV and MERS-CoV are classified as BSL-3 viruses, which makes experimentation into the cellular mechanisms involved in infection more challenging. Here, we use IBV, a BSL-2 virus, as a model for studying the role of Abl kinase activity during coronavirus infection. We found that imatinib and two specific Abl kinase inhibitors, GNF2 and GNF5, reduce IBV titres by blocking the first round of virus infection. Additionally, all three drugs prevented IBV S-induced syncytia formation prior to the hemifusion step. Our results indicate that membrane fusion (both virus-cell and cell-cell) is blocked in the presence of Abl kinase inhibitors. Studying the effects of Abl kinase inhibitors on IBV will be useful in identifying the host cell pathways required for coronavirus infection. This will provide an insight into possible therapeutic targets to treat infections by current as well as newly emerging coronaviruses.
Keywords: Abl kinase; Abl1; Abl2; GNF2; GNF5; IBV; MERS-CoV; SARS-CoV; cell-cell fusion; coronavirus; imatinib; virus-cell fusion.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion.J Virol. 2016 Sep 12;90(19):8924-33. doi: 10.1128/JVI.01429-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27466418 Free PMC article.
-
Identification of Residues Controlling Restriction versus Enhancing Activities of IFITM Proteins on Entry of Human Coronaviruses.J Virol. 2018 Feb 26;92(6):e01535-17. doi: 10.1128/JVI.01535-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263263 Free PMC article.
-
Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation.PLoS One. 2013 Oct 3;8(10):e76469. doi: 10.1371/journal.pone.0076469. eCollection 2013. PLoS One. 2013. PMID: 24098509 Free PMC article.
-
[Development of peptidic MERS-CoV entry inhibitors].Yao Xue Xue Bao. 2015 Dec;50(12):1513-9. Yao Xue Xue Bao. 2015. PMID: 27169270 Review. Chinese.
-
[Cell entry mechanisms of coronaviruses].Uirusu. 2009 Dec;59(2):215-22. doi: 10.2222/jsv.59.215. Uirusu. 2009. PMID: 20218330 Review. Japanese.
Cited by
-
A Review on SARS-CoV-2-Induced Neuroinflammation, Neurodevelopmental Complications, and Recent Updates on the Vaccine Development.Mol Neurobiol. 2021 Sep;58(9):4535-4563. doi: 10.1007/s12035-021-02399-6. Epub 2021 Jun 5. Mol Neurobiol. 2021. PMID: 34089508 Free PMC article. Review.
-
Chronic myeloid leukemia management at the time of the COVID-19 pandemic in Italy. A campus CML survey.Leukemia. 2020 Aug;34(8):2260-2261. doi: 10.1038/s41375-020-0904-z. Epub 2020 Jun 18. Leukemia. 2020. PMID: 32555369 Free PMC article. No abstract available.
-
COVID-19 therapy: What weapons do we bring into battle?Bioorg Med Chem. 2020 Dec 1;28(23):115757. doi: 10.1016/j.bmc.2020.115757. Epub 2020 Sep 10. Bioorg Med Chem. 2020. PMID: 32992245 Free PMC article.
-
Emerging trends from COVID-19 research registered in the Clinical Trials Registry - India.Indian J Med Res. 2021 Jan & Feb;153(1 & 2):26-63. doi: 10.4103/ijmr.IJMR_2556_20. Indian J Med Res. 2021. PMID: 33818466 Free PMC article. Review.
-
Screening of potential inhibitors of COVID-19 with repurposing approach via molecular docking.Netw Model Anal Health Inform Bioinform. 2022;11(1):11. doi: 10.1007/s13721-021-00341-3. Epub 2022 Feb 4. Netw Model Anal Health Inform Bioinform. 2022. PMID: 35136710 Free PMC article.
References
-
- Mingo RM, Simmons JA, Shoemaker CJ, Nelson EA, Schornberg KL, et al. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J Virol. 2015;89:2931–2943. doi: 10.1128/JVI.03398-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

