The Macromolecular Machines that Duplicate the Escherichia coli Chromosome as Targets for Drug Discovery
- PMID: 29538288
- PMCID: PMC5872134
- DOI: 10.3390/antibiotics7010023
The Macromolecular Machines that Duplicate the Escherichia coli Chromosome as Targets for Drug Discovery
Abstract
DNA replication is an essential process. Although the fundamental strategies to duplicate chromosomes are similar in all free-living organisms, the enzymes of the three domains of life that perform similar functions in DNA replication differ in amino acid sequence and their three-dimensional structures. Moreover, the respective proteins generally utilize different enzymatic mechanisms. Hence, the replication proteins that are highly conserved among bacterial species are attractive targets to develop novel antibiotics as the compounds are unlikely to demonstrate off-target effects. For those proteins that differ among bacteria, compounds that are species-specific may be found. Escherichia coli has been developed as a model system to study DNA replication, serving as a benchmark for comparison. This review summarizes the functions of individual E. coli proteins, and the compounds that inhibit them.
Keywords: DNA gyrase; DNA ligase; DNA polymerase I; DNA polymerase III holoenzyme; DNA replication; DnaA; DnaB; DnaC; DnaG; Escherichia coli; SSB; clamp loader; inhibitors; initiation; primase; replication fork; replication origin; replisome; sliding clamp; topoisomerase IV.
Conflict of interest statement
The author expresses no conflict of interest.
Figures
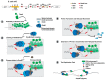
Similar articles
-
Replication Initiation in Bacteria.Enzymes. 2016;39:1-30. doi: 10.1016/bs.enz.2016.03.001. Epub 2016 Apr 20. Enzymes. 2016. PMID: 27241926 Free PMC article. Review.
-
The E. coli DNA Replication Fork.Enzymes. 2016;39:31-88. doi: 10.1016/bs.enz.2016.04.001. Epub 2016 May 13. Enzymes. 2016. PMID: 27241927 Review.
-
Overexpression of the Replicative Helicase in Escherichia coli Inhibits Replication Initiation and Replication Fork Reloading.J Mol Biol. 2016 Mar 27;428(6):1068-1079. doi: 10.1016/j.jmb.2016.01.018. Epub 2016 Jan 23. J Mol Biol. 2016. PMID: 26812209 Free PMC article.
-
Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome.Cell. 1986 Apr 11;45(1):53-64. doi: 10.1016/0092-8674(86)90537-4. Cell. 1986. PMID: 3006926
-
Protein--protein interactions in the eubacterial replisome.IUBMB Life. 2005 Jan;57(1):5-12. doi: 10.1080/15216540500058956. IUBMB Life. 2005. PMID: 16036556 Review.
Cited by
-
Where and When Bacterial Chromosome Replication Starts: A Single Cell Perspective.Front Microbiol. 2018 Nov 26;9:2819. doi: 10.3389/fmicb.2018.02819. eCollection 2018. Front Microbiol. 2018. PMID: 30534115 Free PMC article. Review.
-
SirA inhibits the essential DnaA:DnaD interaction to block helicase recruitment during Bacillus subtilis sporulation.Nucleic Acids Res. 2023 May 22;51(9):4302-4321. doi: 10.1093/nar/gkac1060. Nucleic Acids Res. 2023. PMID: 36416272 Free PMC article.
-
Convergent evolution in two bacterial replicative helicase loaders.Trends Biochem Sci. 2022 Jul;47(7):620-630. doi: 10.1016/j.tibs.2022.02.005. Epub 2022 Mar 26. Trends Biochem Sci. 2022. PMID: 35351361 Free PMC article. Review.
-
Blocking the Trigger: Inhibition of the Initiation of Bacterial Chromosome Replication as an Antimicrobial Strategy.Antibiotics (Basel). 2019 Aug 6;8(3):111. doi: 10.3390/antibiotics8030111. Antibiotics (Basel). 2019. PMID: 31390740 Free PMC article. Review.
-
Quantitative mapping of mRNA 3' ends in Pseudomonas aeruginosa reveals a pervasive role for premature 3' end formation in response to azithromycin.PLoS Genet. 2021 Jul 12;17(7):e1009634. doi: 10.1371/journal.pgen.1009634. eCollection 2021 Jul. PLoS Genet. 2021. PMID: 34252072 Free PMC article.
References
-
- Ear Infections (Otitis Media) in Children (0–17): Use and Expenditures. [(accessed on 15 December 2017)];2006 Available online: https://meps.ahrq.gov/data_files/publications/st228/stat228.pdf.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

