Human Parainfluenza Virus Type 3 Matrix Protein Reduces Viral RNA Synthesis of HPIV3 by Regulating Inclusion Body Formation
- PMID: 29534486
- PMCID: PMC5869518
- DOI: 10.3390/v10030125
Human Parainfluenza Virus Type 3 Matrix Protein Reduces Viral RNA Synthesis of HPIV3 by Regulating Inclusion Body Formation
Abstract
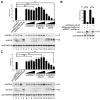
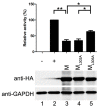
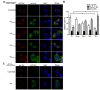
Human parainfluenza virus type 3 is one of the main causes of lower respiratory illness in newborns and infants. The role of the matrix protein (M) in viral budding is extensively studied, but the effect of M on viral replication remains to be determined. Using an HPIV3 minigenome assay, we found that M reduced HPIV3 mingenome-encoded reporter activity even though it had an unspecific effect on the expression of cellular genes. Furthermore, the inhibition effect of M on viral RNA synthesis was proven to be independent of its virus-like particles (VLPs)' release ability. A VLP's defective mutant (ML302A) decreased the expression of minigenome reporter as wild type M did. Using an immunofluorescence assay, we found that M weakened the formation of inclusion bodies (IBs), although it did not co-localize with the IBs. Moreover, using another mutant, ML305A , which is defective in M-nucleoprotein (N) interaction, we found that ML305A had no effect on reporter activity and IB formation as the wild type of M did. Taken together, we conclude that M reduces the replication of HPIV3 and IB formation by M-N interaction.
Keywords: M–N interaction; human parainfluenza virus type 3; inclusion body formation; viral replication; virus-like particles.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Inclusion Body Fusion of Human Parainfluenza Virus Type 3 Regulated by Acetylated α-Tubulin Enhances Viral Replication.J Virol. 2017 Jan 18;91(3):e01802-16. doi: 10.1128/JVI.01802-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881643 Free PMC article.
-
Interaction of Human Parainfluenza Virus Type 3 Nucleoprotein with Matrix Protein Mediates Internal Viral Protein Assembly.J Virol. 2015 Dec 9;90(5):2306-15. doi: 10.1128/JVI.02324-15. J Virol. 2015. PMID: 26656716 Free PMC article.
-
A leucine residue in the C terminus of human parainfluenza virus type 3 matrix protein is essential for efficient virus-like particle and virion release.J Virol. 2014 Nov;88(22):13173-88. doi: 10.1128/JVI.01485-14. Epub 2014 Sep 3. J Virol. 2014. PMID: 25187547 Free PMC article.
-
Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease.J Clin Invest. 2005 Jul;115(7):1688-98. doi: 10.1172/JCI25669. J Clin Invest. 2005. PMID: 16007245 Free PMC article. Review.
-
Structure and Function of Negri Bodies.Adv Exp Med Biol. 2019;1215:111-127. doi: 10.1007/978-3-030-14741-9_6. Adv Exp Med Biol. 2019. PMID: 31317498 Review.
Cited by
-
The two-stage interaction of Ebola virus VP40 with nucleoprotein results in a switch from viral RNA synthesis to virion assembly/budding.Protein Cell. 2022 Feb;13(2):120-140. doi: 10.1007/s13238-020-00764-0. Epub 2020 Nov 3. Protein Cell. 2022. PMID: 33141416 Free PMC article.
-
Isolation, genome analysis and comparison of a novel parainfluenza virus 5 from a Siberian tiger (Panthera tigris).Front Vet Sci. 2024 Apr 5;11:1356378. doi: 10.3389/fvets.2024.1356378. eCollection 2024. Front Vet Sci. 2024. PMID: 38686028 Free PMC article.
-
The W195 Residue of the Newcastle Disease Virus V Protein Is Critical for Multiple Aspects of Viral Self-Regulation through Interactions between V and Nucleoproteins.Viruses. 2024 Apr 10;16(4):584. doi: 10.3390/v16040584. Viruses. 2024. PMID: 38675926 Free PMC article.
-
Human parainfluenza virus 3 vaccine candidates attenuated by codon-pair deoptimization are immunogenic and protective in hamsters.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2316376121. doi: 10.1073/pnas.2316376121. Epub 2024 Jun 11. Proc Natl Acad Sci U S A. 2024. PMID: 38861603 Free PMC article.
-
Curcumin Inhibits Replication of Human Parainfluenza Virus Type 3 by Affecting Viral Inclusion Body Formation.Biomed Res Int. 2021 Aug 9;2021:1807293. doi: 10.1155/2021/1807293. eCollection 2021. Biomed Res Int. 2021. PMID: 34409100 Free PMC article.
References
-
- Longhi S., Receveur-Brechot V., Karlin D., Johansson K., Darbon H., Bhella D., Yeo R., Finet S., Canard B. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 2003;278:18638–18648. doi: 10.1074/jbc.M300518200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

