Neutron Crystallography Detects Differences in Protein Dynamics: Structure of the PKG II Cyclic Nucleotide Binding Domain in Complex with an Activator
- PMID: 29517905
- PMCID: PMC5890435
- DOI: 10.1021/acs.biochem.8b00010
Neutron Crystallography Detects Differences in Protein Dynamics: Structure of the PKG II Cyclic Nucleotide Binding Domain in Complex with an Activator
Abstract
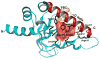
As one of the main receptors of a second messenger, cGMP, cGMP-dependent protein kinase (PKG) isoforms I and II regulate distinct physiological processes. The design of isoform-specific activators is thus of great biomedical importance and requires detailed structural information about PKG isoforms bound with activators, including accurate positions of hydrogen atoms and a description of the hydrogen bonding and water architecture. Here, we determined a 2.2 Å room-temperature joint X-ray/neutron (XN) structure of the human PKG II carboxyl cyclic nucleotide binding (CNB-B) domain bound with a potent PKG II activator, 8-pCPT-cGMP. The XN structure directly visualizes intermolecular interactions and reveals changes in hydrogen bonding patterns upon comparison to the X-ray structure determined at cryo-temperatures. Comparative analysis of the backbone hydrogen/deuterium exchange patterns in PKG II:8-pCPT-cGMP and previously reported PKG Iβ:cGMP XN structures suggests that the ability of these agonists to activate PKG is related to how effectively they quench dynamics of the cyclic nucleotide binding pocket and the surrounding regions.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

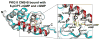

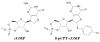
Similar articles
-
Neutron diffraction reveals hydrogen bonds critical for cGMP-selective activation: insights for cGMP-dependent protein kinase agonist design.Biochemistry. 2014 Nov 4;53(43):6725-7. doi: 10.1021/bi501012v. Epub 2014 Oct 22. Biochemistry. 2014. PMID: 25271401 Free PMC article.
-
Structural Basis of Analog Specificity in PKG I and II.ACS Chem Biol. 2017 Sep 15;12(9):2388-2398. doi: 10.1021/acschembio.7b00369. Epub 2017 Aug 22. ACS Chem Biol. 2017. PMID: 28793191 Free PMC article.
-
Crystal structure of cGMP-dependent protein kinase Iβ cyclic nucleotide-binding-B domain : Rp-cGMPS complex reveals an apo-like, inactive conformation.FEBS Lett. 2017 Jan;591(1):221-230. doi: 10.1002/1873-3468.12505. Epub 2016 Dec 23. FEBS Lett. 2017. PMID: 27914169 Free PMC article.
-
Cyclic nucleotide selectivity of protein kinase G isozymes.Protein Sci. 2021 Feb;30(2):316-327. doi: 10.1002/pro.4008. Epub 2020 Dec 10. Protein Sci. 2021. PMID: 33271627 Free PMC article. Review.
-
cAMP-Dependent Protein Kinase and cGMP-Dependent Protein Kinase as Cyclic Nucleotide Effectors.Handb Exp Pharmacol. 2017;238:105-122. doi: 10.1007/164_2015_36. Handb Exp Pharmacol. 2017. PMID: 27885524 Review.
References
-
- Feil R, Lohmann SM, de Jonge H, Walter U, Hofmann F. Cyclic GMP-Dependent Protein Kinase and the Cardiovascular System. Insights from Genetically Modified Mice. Circ Res. 2003;93:907–916. - PubMed
-
- Butt E, Geiger J, Jarchau T, Lohmann SM, Walter U. The cGMP-dependent protein kinase: gene, protein, and function. Neurochem Res. 1993;18:27–42. - PubMed
-
- Hofmann F, Bernhard D, Lukowski R, Weinmeister P. cGMP-regulated protein kinases (cGK) Handb Exp Pharmacol. 2009;191:137–162. - PubMed
-
- Reed RB, Sandberg M, Jahnsen T, Lohmann SM, Francis SH, Corbin JD. Fast and Slow Cyclic Nucleotide-dissociation Sites in cAMP-dependent Protein Kinase Are Transposed in Type 1β cGMP-dependent Protein kinase. J Biol Chem. 1996;271:17570–17575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

