New tools for old drugs: Functional genetic screens to optimize current chemotherapy
- PMID: 29499836
- PMCID: PMC5844649
- DOI: 10.1016/j.drup.2018.01.001
New tools for old drugs: Functional genetic screens to optimize current chemotherapy
Abstract
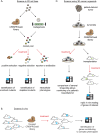
Despite substantial advances in the treatment of various cancers, many patients still receive anti-cancer therapies that hardly eradicate tumor cells but inflict considerable side effects. To provide the best treatment regimen for an individual patient, a major goal in molecular oncology is to identify predictive markers for a personalized therapeutic strategy. Regarding novel targeted anti-cancer therapies, there are usually good markers available. Unfortunately, however, targeted therapies alone often result in rather short remissions and little cytotoxic effect on the cancer cells. Therefore, classical chemotherapy with frequent long remissions, cures, and a clear effect on cancer cell eradication remains a corner stone in current anti-cancer therapy. Reliable biomarkers which predict the response of tumors to classical chemotherapy are rare, in contrast to the situation for targeted therapy. For the bulk of cytotoxic therapeutic agents, including DNA-damaging drugs, drugs targeting microtubules or antimetabolites, there are still no reliable biomarkers used in the clinic to predict tumor response. To make progress in this direction, meticulous studies of classical chemotherapeutic drug action and resistance mechanisms are required. For this purpose, novel functional screening technologies have emerged as successful technologies to study chemotherapeutic drug response in a variety of models. They allow a systematic analysis of genetic contributions to a drug-responsive or -sensitive phenotype and facilitate a better understanding of the mode of action of these drugs. These functional genomic approaches are not only useful for the development of novel targeted anti-cancer drugs but may also guide the use of classical chemotherapeutic drugs by deciphering novel mechanisms influencing a tumor's drug response. Moreover, due to the advances of 3D organoid cultures from patient tumors and in vivo screens in mice, these genetic screens can be applied using conditions that are more representative of the clinical setting. Patient-derived 3D organoid lines furthermore allow the characterization of the "essentialome", the specific set of genes required for survival of these cells, of an individual tumor, which could be monitored over the course of treatment and help understanding how drug resistance evolves in clinical tumors. Thus, we expect that these functional screens will enable the discovery of novel cancer-specific vulnerabilities, and through clinical validation, move the field of predictive biomarkers forward. This review focuses on novel advanced techniques to decipher the interplay between genetic alterations and drug response.
Keywords: 3D organoids; CRISPR/Cas9; Chemotherapy; DNA damage; Functional genetic screens; Gene essentiality; Haploid cells; Insertional mutagenesis; Predictive markers.
Copyright © 2018 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
Functional-genetic approaches to understanding drug response and resistance.Curr Opin Genet Dev. 2019 Feb;54:41-47. doi: 10.1016/j.gde.2019.03.003. Epub 2019 Apr 2. Curr Opin Genet Dev. 2019. PMID: 30951975 Review.
-
Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response.JAMA Oncol. 2015 Jul;1(4):466-74. doi: 10.1001/jamaoncol.2015.1313. JAMA Oncol. 2015. PMID: 26181256 Free PMC article.
-
CRISPR screen in mechanism and target discovery for cancer immunotherapy.Biochim Biophys Acta Rev Cancer. 2020 Aug;1874(1):188378. doi: 10.1016/j.bbcan.2020.188378. Epub 2020 May 13. Biochim Biophys Acta Rev Cancer. 2020. PMID: 32413572 Review.
-
Agreement between two large pan-cancer CRISPR-Cas9 gene dependency data sets.Nat Commun. 2019 Dec 20;10(1):5817. doi: 10.1038/s41467-019-13805-y. Nat Commun. 2019. PMID: 31862961 Free PMC article.
-
Circulating tumor cell-derived organoids: Current challenges and promises in medical research and precision medicine.Biochim Biophys Acta Rev Cancer. 2018 Apr;1869(2):117-127. doi: 10.1016/j.bbcan.2017.12.005. Epub 2018 Jan 31. Biochim Biophys Acta Rev Cancer. 2018. PMID: 29360544 Free PMC article. Review.
Cited by
-
Genome-wide CRISPR screens of oral squamous cell carcinoma reveal fitness genes in the Hippo pathway.Elife. 2020 Sep 29;9:e57761. doi: 10.7554/eLife.57761. Elife. 2020. PMID: 32990596 Free PMC article.
-
CRISPR screens guide the way for PARP and ATR inhibitor biomarker discovery.FEBS J. 2022 Dec;289(24):7854-7868. doi: 10.1111/febs.16217. Epub 2021 Oct 17. FEBS J. 2022. PMID: 34601817 Free PMC article. Review.
-
Global cluster analysis and network visualization in organoids in cancer research: a scientometric mapping from 1991 to 2021.Front Oncol. 2023 Sep 15;13:1253573. doi: 10.3389/fonc.2023.1253573. eCollection 2023. Front Oncol. 2023. PMID: 37781203 Free PMC article.
-
Pooled Screens Identify GPR108 and TM9SF2 as Host Cell Factors Critical for AAV Transduction.Mol Ther Methods Clin Dev. 2020 Mar 17;17:601-611. doi: 10.1016/j.omtm.2020.03.012. eCollection 2020 Jun 12. Mol Ther Methods Clin Dev. 2020. PMID: 32280726 Free PMC article.
-
Applications of Cancer Cell-Specific Aptamers in Targeted Delivery of Anticancer Therapeutic Agents.Molecules. 2018 Apr 4;23(4):830. doi: 10.3390/molecules23040830. Molecules. 2018. PMID: 29617327 Free PMC article. Review.
References
-
- Aggarwal S. Targeted cancer therapies. Nat. Rev. Drug Discov. 2010;9:427–428. - PubMed
-
- Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B., Kost-Alimova M., Gill S., Xu H., Ali L.D., Jiang G.Z., Pantel S., Lee Y., Goodale A., Cherniack A.D., Oh C., Kryukov G., Cowley G.S., Garraway L.A., Stegmaier K., Roberts C.W., Golub T.R., Meyerson M., Root D.E., Tsherniak A., Hahn W.C. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016;6:914–929. - PMC - PubMed
-
- Al-Lazikani B., Banerji U., Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012;30:679. - PubMed
-
- Annunziato S., Kas S.M., Nethe M., Yucel H., Del Bravo J., Pritchard C., Bin Ali R., Van Gerwen B., Siteur B., Drenth A.P., Schut E., Van De Ven M., Boelens M.C., Klarenbeek S., Huijbers I.J., Van Miltenburg M.H., Jonkers J. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes. Dev. 2016;30:1470–1480. - PMC - PubMed
-
- Babij C., Zhang Y.H., Kurzeja R.J., Munzli A., Shehabeldin A., Fernando M., Quon K., Kassner P.D., Ruefli-Brasse A.A., Watson V.J., Fajardo F., Jackson A., Zondlo J., Sun Y., Ellison A.R., Plewa C.A., San Miguel T., Robinson J., Mccarter J., Schwandner R., Judd T., Carnahan J., Dussault I. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71:5818–5826. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

