Nubbin isoform antagonism governs Drosophila intestinal immune homeostasis
- PMID: 29499056
- PMCID: PMC5851638
- DOI: 10.1371/journal.ppat.1006936
Nubbin isoform antagonism governs Drosophila intestinal immune homeostasis
Abstract
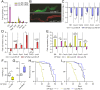
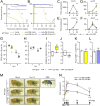
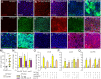
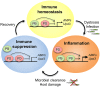
Gut immunity is regulated by intricate and dynamic mechanisms to ensure homeostasis despite a constantly changing microbial environment. Several regulatory factors have been described to participate in feedback responses to prevent aberrant immune activity. Little is, however, known about how transcriptional programs are directly tuned to efficiently adapt host gut tissues to the current microbiome. Here we show that the POU/Oct gene nubbin (nub) encodes two transcription factor isoforms, Nub-PB and Nub-PD, which antagonistically regulate immune gene expression in Drosophila. Global transcriptional profiling of adult flies overexpressing Nub-PB in immunocompetent tissues revealed that this form is a strong transcriptional activator of a large set of immune genes. Further genetic analyses showed that Nub-PB is sufficient to drive expression both independently and in conjunction with nuclear factor kappa B (NF-κB), JNK and JAK/STAT pathways. Similar overexpression of Nub-PD did, conversely, repress expression of the same targets. Strikingly, isoform co-overexpression normalized immune gene transcription, suggesting antagonistic activities. RNAi-mediated knockdown of individual nub transcripts in enterocytes confirmed antagonistic regulation by the two isoforms and that both are necessary for normal immune gene transcription in the midgut. Furthermore, enterocyte-specific Nub-PB expression levels had a strong impact on gut bacterial load as well as host lifespan. Overexpression of Nub-PB enhanced bacterial clearance of ingested Erwinia carotovora carotovora 15. Nevertheless, flies quickly succumbed to the infection, suggesting a deleterious immune response. In line with this, prolonged overexpression promoted a proinflammatory signature in the gut with induction of JNK and JAK/STAT pathways, increased apoptosis and stem cell proliferation. These findings highlight a novel regulatory mechanism of host-microbe interactions mediated by antagonistic transcription factor isoforms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The POU/Oct Transcription Factor Nubbin Controls the Balance of Intestinal Stem Cell Maintenance and Differentiation by Isoform-Specific Regulation.Stem Cell Reports. 2018 May 8;10(5):1565-1578. doi: 10.1016/j.stemcr.2018.03.014. Epub 2018 Apr 19. Stem Cell Reports. 2018. PMID: 29681543 Free PMC article.
-
The Oct1 homolog Nubbin is a repressor of NF-κB-dependent immune gene expression that increases the tolerance to gut microbiota.BMC Biol. 2013 Sep 6;11:99. doi: 10.1186/1741-7007-11-99. BMC Biol. 2013. PMID: 24010524 Free PMC article.
-
The POU/Oct Transcription Factor Pdm1/nub Is Necessary for a Beneficial Gut Microbiota and Normal Lifespan of Drosophila.J Innate Immun. 2016;8(4):412-26. doi: 10.1159/000446368. Epub 2016 May 28. J Innate Immun. 2016. PMID: 27231014 Free PMC article.
-
Regulation of immune and tissue homeostasis by Drosophila POU factors.Insect Biochem Mol Biol. 2019 Jun;109:24-30. doi: 10.1016/j.ibmb.2019.04.003. Epub 2019 Apr 5. Insect Biochem Mol Biol. 2019. PMID: 30954681 Review.
-
Microbial Control of Intestinal Homeostasis via Enteroendocrine Cell Innate Immune Signaling.Trends Microbiol. 2020 Feb;28(2):141-149. doi: 10.1016/j.tim.2019.09.005. Epub 2019 Nov 4. Trends Microbiol. 2020. PMID: 31699645 Free PMC article. Review.
Cited by
-
HMGB1-Like Dorsal Switch Protein 1 Triggers a Damage Signal in Mosquito Gut to Activate Dual Oxidase via Eicosanoids.J Innate Immun. 2022;14(6):657-672. doi: 10.1159/000524561. Epub 2022 May 5. J Innate Immun. 2022. PMID: 35512659 Free PMC article.
-
Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster.Genetics. 2018 Oct;210(2):357-396. doi: 10.1534/genetics.118.300224. Genetics. 2018. PMID: 30287514 Free PMC article. Review.
-
The POU/Oct Transcription Factor Nubbin Controls the Balance of Intestinal Stem Cell Maintenance and Differentiation by Isoform-Specific Regulation.Stem Cell Reports. 2018 May 8;10(5):1565-1578. doi: 10.1016/j.stemcr.2018.03.014. Epub 2018 Apr 19. Stem Cell Reports. 2018. PMID: 29681543 Free PMC article.
-
JNK Signaling in Drosophila Aging and Longevity.Int J Mol Sci. 2021 Sep 6;22(17):9649. doi: 10.3390/ijms22179649. Int J Mol Sci. 2021. PMID: 34502551 Free PMC article. Review.
-
Stop codon readthrough alters the activity of a POU/Oct transcription factor during Drosophila development.BMC Biol. 2021 Sep 3;19(1):185. doi: 10.1186/s12915-021-01106-0. BMC Biol. 2021. PMID: 34479564 Free PMC article.
References
-
- Bergman P, Seyedoleslami Esfahani S, Engstrom Y. Drosophila as a Model for Human Diseases-Focus on Innate Immunity in Barrier Epithelia. Curr Top Dev Biol. 2017;121:29–81. doi: 10.1016/bs.ctdb.2016.07.002 - DOI - PubMed
-
- Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster—from microbial recognition to whole-organism physiology. Nat Rev Immunol. 2014;14(12):796–810. doi: 10.1038/nri3763 - DOI - PMC - PubMed
-
- Lindsay SA, Wasserman SA. Conventional and non-conventional Drosophila Toll signaling. Dev Comp Immunol. 2014;42(1):16–24. doi: 10.1016/j.dci.2013.04.011 - DOI - PMC - PubMed
-
- Liu X, Hodgson JJ, Buchon N. Drosophila as a model for homeostatic, antibacterial, and antiviral mechanisms in the gut. PLoS Pathog. 2017;13(5):e1006277 doi: 10.1371/journal.ppat.1006277 - DOI - PMC - PubMed
-
- Mansour SC, Pena OM, Hancock RE. Host defense peptides: front-line immunomodulators. Trends Immunol. 2014;35(9):443–50. doi: 10.1016/j.it.2014.07.004 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

