Biochemical characterization and essentiality of Plasmodium fumarate hydratase
- PMID: 29449371
- PMCID: PMC5912476
- DOI: 10.1074/jbc.M117.816298
Biochemical characterization and essentiality of Plasmodium fumarate hydratase
Abstract
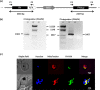
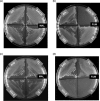
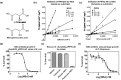
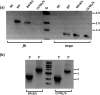
Plasmodium falciparum (Pf), the causative agent of malaria, has an iron-sulfur cluster-containing class I fumarate hydratase (FH) that catalyzes the interconversion of fumarate to malate, a well-known reaction in the tricarboxylic acid cycle. In humans, the same reaction is catalyzed by class II FH that has no sequence or structural homology with the class I enzyme from Plasmodium Fumarate is generated in large quantities in the parasite as a by-product of AMP synthesis and is converted to malate by FH and then used in the generation of the key metabolites oxaloacetate, aspartate, and pyruvate. Previous studies have identified the FH reaction as being essential to P. falciparum, but biochemical characterization of PfFH that may provide leads for the development of specific inhibitors is lacking. Here, we report on the kinetic characterization of purified recombinant PfFH, functional complementation of fh deficiency in Escherichia coli, and mitochondrial localization in the parasite. We found that the substrate analog mercaptosuccinic acid is a potent PfFH inhibitor, with a Ki value in the nanomolar range. The fh gene could not be knocked out in Plasmodium berghei when transfectants were introduced into BALB/c mice; however, fh knockout was successful when C57BL/6 mice were used as host, suggesting that the essentiality of the fh gene to the parasite was mouse strain-dependent.
Keywords: Plasmodium; class I fumarate hydratase; enzyme inhibitor; essentiality of fumarate hydratase; gene knockout; mercaptosuccinic acid; parasitology; tricarboxylic acid cycle (TCA cycle) (Krebs cycle).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Metabolic changes accompanying the loss of fumarate hydratase and malate-quinone oxidoreductase in the asexual blood stage of Plasmodium falciparum.J Biol Chem. 2022 May;298(5):101897. doi: 10.1016/j.jbc.2022.101897. Epub 2022 Apr 6. J Biol Chem. 2022. PMID: 35398098 Free PMC article.
-
Suppression of experimental cerebral malaria by disruption of malate:quinone oxidoreductase.Malar J. 2017 Jun 12;16(1):247. doi: 10.1186/s12936-017-1898-5. Malar J. 2017. PMID: 28606087 Free PMC article.
-
Metabolic fate of fumarate, a side product of the purine salvage pathway in the intraerythrocytic stages of Plasmodium falciparum.J Biol Chem. 2011 Mar 18;286(11):9236-45. doi: 10.1074/jbc.M110.173328. Epub 2011 Jan 5. J Biol Chem. 2011. PMID: 21209090 Free PMC article.
-
Functional implications of fumarate-induced cysteine succination.Trends Biochem Sci. 2024 Sep;49(9):775-790. doi: 10.1016/j.tibs.2024.05.003. Epub 2024 Jun 13. Trends Biochem Sci. 2024. PMID: 38876954 Review.
-
The role of cGMP signalling in regulating life cycle progression of Plasmodium.Microbes Infect. 2012 Aug;14(10):831-7. doi: 10.1016/j.micinf.2012.04.011. Epub 2012 May 3. Microbes Infect. 2012. PMID: 22613210 Free PMC article. Review.
Cited by
-
Interchangeability of class I and II fumarases in an obligate methanotroph Methylotuvimicrobium alcaliphilum 20Z.PLoS One. 2023 Oct 26;18(10):e0289976. doi: 10.1371/journal.pone.0289976. eCollection 2023. PLoS One. 2023. PMID: 37883386 Free PMC article.
-
Metabolic changes accompanying the loss of fumarate hydratase and malate-quinone oxidoreductase in the asexual blood stage of Plasmodium falciparum.J Biol Chem. 2022 May;298(5):101897. doi: 10.1016/j.jbc.2022.101897. Epub 2022 Apr 6. J Biol Chem. 2022. PMID: 35398098 Free PMC article.
-
Structural and Biochemical Investigations of the [4Fe-4S] Cluster-Containing Fumarate Hydratase from Leishmania major.Biochemistry. 2019 Dec 10;58(49):5011-5021. doi: 10.1021/acs.biochem.9b00923. Epub 2019 Nov 27. Biochemistry. 2019. PMID: 31743022 Free PMC article.
-
Crystal Structures of Fumarate Hydratases from Leishmania major in a Complex with Inhibitor 2-Thiomalate.ACS Chem Biol. 2019 Feb 15;14(2):266-275. doi: 10.1021/acschembio.8b00972. Epub 2019 Jan 24. ACS Chem Biol. 2019. PMID: 30645090 Free PMC article.
-
Divergent acyl carrier protein decouples mitochondrial Fe-S cluster biogenesis from fatty acid synthesis in malaria parasites.Elife. 2021 Oct 6;10:e71636. doi: 10.7554/eLife.71636. Elife. 2021. PMID: 34612205 Free PMC article.
References
-
- Roth E. (1990) Plasmodium falciparum carbohydrate metabolism: a connection between host cell and parasite. Blood Cells 16, 453–460 - PubMed
-
- MacRae J. I., Dixon M. W., Dearnley M. K., Chua H. H., Chambers J. M., Kenny S., Bottova I., Tilley L., and McConville M. J. (2013) Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol. 11, 67 10.1186/1741-7007-11-67 - DOI - PMC - PubMed
-
- Teipel J. W., Hass G. M., and Hill R. L. (1968) The substrate specificity of fumarase. J. Biol. Chem. 243, 5684–5694 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

