FVIII proteins with a modified immunodominant T-cell epitope exhibit reduced immunogenicity and normal FVIII activity
- PMID: 29444872
- PMCID: PMC5858479
- DOI: 10.1182/bloodadvances.2017013482
FVIII proteins with a modified immunodominant T-cell epitope exhibit reduced immunogenicity and normal FVIII activity
Abstract
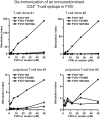
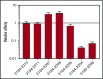
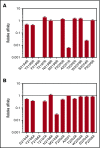
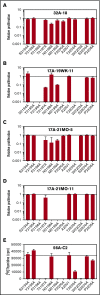
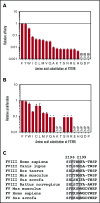
Factor VIII (FVIII)-neutralizing antibodies (inhibitors) are a serious complication in hemophilia A (HA). The peptide FVIII2194-2213 contains an immunodominant HLA-DRA*01-DRB1*01:01 (DRB1*01:01)-restricted epitope recognized by CD4+ T-effector cells from HA subjects. The aim of this study was to identify amino acid substitutions to deimmunize this epitope while retaining procoagulant function and expression levels comparable to those of wild-type (WT) FVIII proteins. The shortest DRB1*01:01-binding peptide was FVIII2194-2205, and residues important for affinity were identified as F2196, M2199, A2201, and S2204. T-cell proliferation experiments with Ala-substituted FVIII2194-2205 peptides identified F2196A as a substitution that abrogated proliferation of clones specific for the WT sequence. T-cell clones that were stimulated by recombinant WT-FVIII-C2 (rWT-FVIII-C2) protein did not proliferate when cultured with rFVIII-C2-F2196A, indicating the immunogenic peptide includes a naturally processed T-cell epitope. Additional amino acid substitutions at F2196 and M2199 were evaluated by peptide-MHC class II (MHCII)-binding assays, T-cell proliferation assays, epitope prediction algorithms, and sequence homologies. Six B-domain-deleted (BDD)-FVIII proteins with substitutions F2196A, F2196L, F2196K, M2199A, M2199W, or M2199R were produced. Proliferation of T-cell clones and polyclonal lines in response to rBDD-FVIII-F2196K and rBDD-FVIII-M2199A was reduced compared with responses to WT-BDD-FVIII. The BDD-FVIII-F2196K sequence modification appears to be the most promising sequence variant tested here, due to its effectiveness at eliminating DRB1*01:01-restricted immunogenicity, low potential immunogenicity in the context of other MHCII alleles, expression level comparable to WT-BDD-FVIII, and retained procoagulant activity. These results provide proof of principle for the design of less immunogenic FVIII proteins targeted to specific subsets of HA patients.
Conflict of interest statement
Conflict-of-interest disclosure: K.P.P., R.A.E., and E.A.J. are inventors on FVIII patents. J.A.L. is currently an employee of Shire. The remaining authors declare no competing financial interests.
Figures
Similar articles
-
Six amino acid residues in a 1200 Å2 interface mediate binding of factor VIII to an IgG4κ inhibitory antibody.PLoS One. 2015 Jan 23;10(1):e0116577. doi: 10.1371/journal.pone.0116577. eCollection 2015. PLoS One. 2015. PMID: 25615825 Free PMC article.
-
T cells from hemophilia A subjects recognize the same HLA-restricted FVIII epitope with a narrow TCR repertoire.Blood. 2016 Oct 20;128(16):2043-2054. doi: 10.1182/blood-2015-11-682468. Epub 2016 Jul 28. Blood. 2016. PMID: 27471234 Free PMC article. Clinical Trial.
-
T-cell responses over time in a mild hemophilia A inhibitor subject: epitope identification and transient immunogenicity of the corresponding self-peptide.J Thromb Haemost. 2007 Dec;5(12):2399-407. doi: 10.1111/j.1538-7836.2007.02762.x. J Thromb Haemost. 2007. PMID: 18034765
-
Hunting down factor VIII in the immunopeptidome.Cell Immunol. 2016 Mar;301:59-64. doi: 10.1016/j.cellimm.2015.11.001. Epub 2015 Nov 5. Cell Immunol. 2016. PMID: 26610639 Review.
-
B-cell and T-cell epitopes in anti-factor VIII immune responses.Clin Rev Allergy Immunol. 2009 Oct;37(2):80-95. doi: 10.1007/s12016-009-8120-7. Clin Rev Allergy Immunol. 2009. PMID: 19184559 Review.
Cited by
-
Immunogenicity Challenges Associated with Subcutaneous Delivery of Therapeutic Proteins.BioDrugs. 2021 Mar;35(2):125-146. doi: 10.1007/s40259-020-00465-4. Epub 2021 Feb 1. BioDrugs. 2021. PMID: 33523413 Free PMC article. Review.
-
The self-reactive FVIII T cell repertoire in healthy individuals relies on a short set of epitopes and public clonotypes.Front Immunol. 2024 Mar 6;15:1345195. doi: 10.3389/fimmu.2024.1345195. eCollection 2024. Front Immunol. 2024. PMID: 38510258 Free PMC article.
-
Treatment of Hemophilia A Using Factor VIII Messenger RNA Lipid Nanoparticles.Mol Ther Nucleic Acids. 2020 Jun 5;20:534-544. doi: 10.1016/j.omtn.2020.03.015. Epub 2020 Apr 7. Mol Ther Nucleic Acids. 2020. PMID: 32330871 Free PMC article.
-
Immunogenicity of Protein Pharmaceuticals.J Pharm Sci. 2019 May;108(5):1637-1654. doi: 10.1016/j.xphs.2018.12.014. Epub 2018 Dec 30. J Pharm Sci. 2019. PMID: 30599169 Free PMC article. Review.
-
Hemophilia A subjects with an intron-22 gene inversion mutation show CD4+ T-effector responses to multiple epitopes in FVIII.Front Immunol. 2023 Mar 1;14:1128641. doi: 10.3389/fimmu.2023.1128641. eCollection 2023. Front Immunol. 2023. PMID: 36936969 Free PMC article.
References
-
- Konkle BA, Huston H, Nakaya Fletcher S. Hemophilia A. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews. Seattle, WA: University of Washington; 2000.
-
- Srivastava A, Brewer AK, Mauser-Bunschoten EP, et al. ; Treatment Guidelines Working Group on Behalf of The World Federation Of Hemophilia. Guidelines for the management of hemophilia. Haemophilia. 2013;19(1):e1-e47. - PubMed
-
- Gouw SC, van den Berg HM, Oldenburg J, et al. . F8 gene mutation type and inhibitor development in patients with severe hemophilia A: systematic review and meta-analysis. Blood. 2012;119(12):2922-2934. - PubMed
-
- Kempton CL, Meeks SL. Toward optimal therapy for inhibitors in hemophilia. Blood. 2014;124(23):3365-3372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

