How many human proteoforms are there?
- PMID: 29443976
- PMCID: PMC5837046
- DOI: 10.1038/nchembio.2576
How many human proteoforms are there?
Abstract
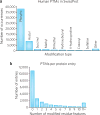
Despite decades of accumulated knowledge about proteins and their post-translational modifications (PTMs), numerous questions remain regarding their molecular composition and biological function. One of the most fundamental queries is the extent to which the combinations of DNA-, RNA- and PTM-level variations explode the complexity of the human proteome. Here, we outline what we know from current databases and measurement strategies including mass spectrometry-based proteomics. In doing so, we examine prevailing notions about the number of modifications displayed on human proteins and how they combine to generate the protein diversity underlying health and disease. We frame central issues regarding determination of protein-level variation and PTMs, including some paradoxes present in the field today. We use this framework to assess existing data and to ask the question, "How many distinct primary structures of proteins (proteoforms) are created from the 20,300 human genes?" We also explore prospects for improving measurements to better regularize protein-level biology and efficiently associate PTMs to function and phenotype.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Mapping intact protein isoforms in discovery mode using top-down proteomics.Nature. 2011 Oct 30;480(7376):254-8. doi: 10.1038/nature10575. Nature. 2011. PMID: 22037311 Free PMC article.
-
Constructing Human Proteoform Families Using Intact-Mass and Top-Down Proteomics with a Multi-Protease Global Post-Translational Modification Discovery Database.J Proteome Res. 2019 Oct 4;18(10):3671-3680. doi: 10.1021/acs.jproteome.9b00339. Epub 2019 Sep 18. J Proteome Res. 2019. PMID: 31479276 Free PMC article.
-
Mass spectrometry-based proteomics analyses of post-translational modifications and proteoforms in human pituitary adenomas.Biochim Biophys Acta Proteins Proteom. 2021 Mar;1869(3):140584. doi: 10.1016/j.bbapap.2020.140584. Epub 2020 Dec 13. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 33321259 Review.
-
The Methods Employed in Mass Spectrometric Analysis of Posttranslational Modifications (PTMs) and Protein-Protein Interactions (PPIs).Adv Exp Med Biol. 2019;1140:169-198. doi: 10.1007/978-3-030-15950-4_10. Adv Exp Med Biol. 2019. PMID: 31347048 Free PMC article. Review.
-
The ubiquitin proteoform problem.Curr Opin Chem Biol. 2021 Aug;63:95-104. doi: 10.1016/j.cbpa.2021.02.015. Epub 2021 Apr 1. Curr Opin Chem Biol. 2021. PMID: 33813043 Free PMC article. Review.
Cited by
-
Observations from the Proteomics Bench.Proteomes. 2024 Feb 6;12(1):6. doi: 10.3390/proteomes12010006. Proteomes. 2024. PMID: 38390966 Free PMC article.
-
mRNAs, proteins and the emerging principles of gene expression control.Nat Rev Genet. 2020 Oct;21(10):630-644. doi: 10.1038/s41576-020-0258-4. Epub 2020 Jul 24. Nat Rev Genet. 2020. PMID: 32709985 Review.
-
hnRNPs: roles in neurodevelopment and implication for brain disorders.Front Mol Neurosci. 2024 Jul 17;17:1411639. doi: 10.3389/fnmol.2024.1411639. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39086926 Free PMC article. Review.
-
Chemical Synthesis of Human Proteoforms and Application in Biomedicine.ACS Cent Sci. 2024 Jul 22;10(8):1442-1459. doi: 10.1021/acscentsci.4c00642. eCollection 2024 Aug 28. ACS Cent Sci. 2024. PMID: 39220697 Free PMC article. Review.
-
Spatiotemporal changes in the human lens proteome: Critical insights into long-lived proteins.Prog Retin Eye Res. 2020 May;76:100802. doi: 10.1016/j.preteyeres.2019.100802. Epub 2019 Nov 6. Prog Retin Eye Res. 2020. PMID: 31704338 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA196539/CA/NCI NIH HHS/United States
- P41 GM108569/GM/NIGMS NIH HHS/United States
- R01 GM114292/GM/NIGMS NIH HHS/United States
- R01 HL111362/HL/NHLBI NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
- P01 HL112730/HL/NHLBI NIH HHS/United States
- R01 HL132075/HL/NHLBI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- S10 OD019938/OD/NIH HHS/United States
- R01 AI118891/AI/NIAID NIH HHS/United States
- R01 GM110174/GM/NIGMS NIH HHS/United States
- R01 GM115739/GM/NIGMS NIH HHS/United States
- R01 GM021248/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

