Dietary Modulation of the Epigenome
- PMID: 29442595
- PMCID: PMC5966714
- DOI: 10.1152/physrev.00010.2017
Dietary Modulation of the Epigenome
Abstract
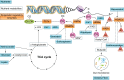
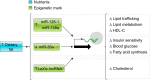
Epigenetics is the study of heritable mechanisms that can modify gene activity and phenotype without modifying the genetic code. The basis for the concept of epigenetics originated more than 2,000 yr ago as a theory to explain organismal development. However, the definition of epigenetics continues to evolve as we identify more of the components that make up the epigenome and dissect the complex manner by which they regulate and are regulated by cellular functions. A substantial and growing body of research shows that nutrition plays a significant role in regulating the epigenome. Here, we critically assess this diverse body of evidence elucidating the role of nutrition in modulating the epigenome and summarize the impact such changes have on molecular and physiological outcomes with regards to human health.
Figures
Similar articles
-
The interaction between epigenetics, nutrition and the development of cancer.Nutrients. 2015 Jan 30;7(2):922-47. doi: 10.3390/nu7020922. Nutrients. 2015. PMID: 25647662 Free PMC article. Review.
-
Nutritional modulation of the epigenome and its implication for future health.Proc Nutr Soc. 2019 Aug;78(3):305-312. doi: 10.1017/S0029665119000016. Epub 2019 Feb 19. Proc Nutr Soc. 2019. PMID: 30777143 Review.
-
Session 2: Personalised nutrition. Epigenomics: a basis for understanding individual differences?Proc Nutr Soc. 2008 Nov;67(4):390-4. doi: 10.1017/S0029665108008744. Proc Nutr Soc. 2008. PMID: 18847515 Review.
-
You are what you eat - How nutrition and metabolism shape the genome through epigenetics.Mol Metab. 2020 Aug;38:100987. doi: 10.1016/j.molmet.2020.100987. Epub 2020 Apr 15. Mol Metab. 2020. PMID: 32314738 Free PMC article. No abstract available.
-
What do studies of insect polyphenisms tell us about nutritionally-triggered epigenomic changes and their consequences?Nutrients. 2015 Mar 11;7(3):1787-97. doi: 10.3390/nu7031787. Nutrients. 2015. PMID: 25768950 Free PMC article. Review.
Cited by
-
Impacts of Epigenetic Processes on the Health and Productivity of Livestock.Front Genet. 2021 Feb 23;11:613636. doi: 10.3389/fgene.2020.613636. eCollection 2020. Front Genet. 2021. PMID: 33708235 Free PMC article. Review.
-
The use of race terms in epigenetics research: considerations moving forward.Front Genet. 2024 Jan 31;15:1348855. doi: 10.3389/fgene.2024.1348855. eCollection 2024. Front Genet. 2024. PMID: 38356697 Free PMC article.
-
(-)-Methyl-Oleocanthal, a New Oleocanthal Metabolite Reduces LPS-Induced Inflammatory and Oxidative Response: Molecular Signaling Pathways and Histones Epigenetic Modulation.Antioxidants (Basel). 2021 Dec 27;11(1):56. doi: 10.3390/antiox11010056. Antioxidants (Basel). 2021. PMID: 35052558 Free PMC article.
-
Biomarkers in critical care nutrition.Crit Care. 2020 Aug 12;24(1):499. doi: 10.1186/s13054-020-03208-7. Crit Care. 2020. PMID: 32787899 Free PMC article. Review.
-
Birthweight predicts glomerular filtration rate in adult-life: population based cross sectional study.Ren Fail. 2021 Dec;43(1):664-675. doi: 10.1080/0886022X.2021.1915798. Ren Fail. 2021. Retraction in: Ren Fail. 2022 Dec;44(1):1622. doi: 10.1080/0886022X.2022.2075579 PMID: 33896360 Free PMC article. Retracted.
References
-
- Amarasekera M, Martino D, Ashley S, Harb H, Kesper D, Strickland D, Saffery R, Prescott SL. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. FASEB J 28: 4068–4076, 2014. doi:10.1096/fj.13-249029. - DOI - PMC - PubMed
-
- Anderson JG, Ramadori G, Ioris RM, Galiè M, Berglund ED, Coate KC, Fujikawa T, Pucciarelli S, Moreschini B, Amici A, Andreani C, Coppari R. Enhanced insulin sensitivity in skeletal muscle and liver by physiological overexpression of SIRT6. Mol Metab 4: 846–856, 2015. doi:10.1016/j.molmet.2015.09.003. - DOI - PMC - PubMed
-
- Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen AS, Green MF, Sivley RM, Ilkayeva OR, Stevens RD, Backos DS, Capra JA, Olsen CA, Campbell JE, Muoio DM, Grimsrud PA, Hirschey MD. SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion. Cell Metab 25: 838–855.e15, 2017. doi:10.1016/j.cmet.2017.03.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

