Asp1 Bifunctional Activity Modulates Spindle Function via Controlling Cellular Inositol Pyrophosphate Levels in Schizosaccharomyces pombe
- PMID: 29440310
- PMCID: PMC5902593
- DOI: 10.1128/MCB.00047-18
Asp1 Bifunctional Activity Modulates Spindle Function via Controlling Cellular Inositol Pyrophosphate Levels in Schizosaccharomyces pombe
Abstract
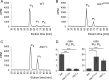
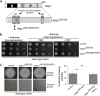
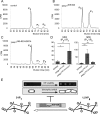
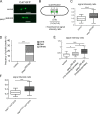
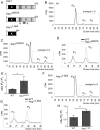
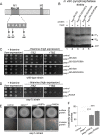
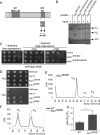
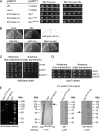
The generation of two daughter cells with the same genetic information requires error-free chromosome segregation during mitosis. Chromosome transmission fidelity is dependent on spindle structure/function, which requires Asp1 in the fission yeast Schizosaccharomyces pombe Asp1 belongs to the diphosphoinositol pentakisphosphate kinase (PPIP5K)/Vip1 family which generates high-energy inositol pyrophosphate (IPP) molecules. Here, we show that Asp1 is a bifunctional enzyme in vivo: Asp1 kinase generates specific IPPs which are the substrates of the Asp1 pyrophosphatase. Intracellular levels of these IPPs directly correlate with microtubule stability: pyrophosphatase loss-of-function mutants raised Asp1-made IPP levels 2-fold, thus increasing microtubule stability, while overexpression of the pyrophosphatase decreased microtubule stability. Absence of Asp1-generated IPPs resulted in an aberrant, increased spindle association of the S. pombe kinesin-5 family member Cut7, which led to spindle collapse. Thus, chromosome transmission is controlled via intracellular IPP levels. Intriguingly, identification of the mitochondrion-associated Met10 protein as the first pyrophosphatase inhibitor revealed that IPPs also regulate mitochondrial distribution.
Keywords: PPIP5K family; Schizosaccharomyces pombe; chromosome segregation; inositol pyrophosphate; microtubule; mitosis; phosphatase; signaling molecules; yeast.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
Inositol Pyrophosphate Kinase Asp1 Modulates Chromosome Segregation Fidelity and Spindle Function in Schizosaccharomyces pombe.Mol Cell Biol. 2016 Nov 28;36(24):3128-3140. doi: 10.1128/MCB.00330-16. Print 2016 Dec 15. Mol Cell Biol. 2016. PMID: 27697865 Free PMC article.
-
Activities and Structure-Function Analysis of Fission Yeast Inositol Pyrophosphate (IPP) Kinase-Pyrophosphatase Asp1 and Its Impact on Regulation of pho1 Gene Expression.mBio. 2022 Jun 28;13(3):e0103422. doi: 10.1128/mbio.01034-22. Epub 2022 May 10. mBio. 2022. PMID: 35536002 Free PMC article.
-
The Asp1 pyrophosphatase from S. pombe hosts a [2Fe-2S]2+ cluster in vivo.J Biol Inorg Chem. 2021 Feb;26(1):93-108. doi: 10.1007/s00775-020-01840-w. Epub 2021 Feb 5. J Biol Inorg Chem. 2021. PMID: 33544225 Free PMC article.
-
Inositol pyrophosphates get the vip1 treatment.Cell. 2007 May 18;129(4):647-9. doi: 10.1016/j.cell.2007.05.002. Cell. 2007. PMID: 17512396 Review.
-
Spatiotemporal control of spindle disassembly in fission yeast.Cell Mol Life Sci. 2019 Sep;76(18):3543-3551. doi: 10.1007/s00018-019-03139-9. Epub 2019 May 25. Cell Mol Life Sci. 2019. PMID: 31129857 Free PMC article. Review.
Cited by
-
Inactivation of fission yeast Erh1 de-represses pho1 expression: evidence that Erh1 is a negative regulator of prt lncRNA termination.RNA. 2020 Oct;26(10):1334-1344. doi: 10.1261/rna.076463.120. Epub 2020 Jun 16. RNA. 2020. PMID: 32546512 Free PMC article.
-
Genetic suppressor screen identifies Tgp1 (glycerophosphocholine transporter), Kcs1 (IP6 kinase), and Plc1 (phospholipase C) as determinants of inositol pyrophosphate toxicosis in fission yeast.mBio. 2024 Feb 14;15(2):e0306223. doi: 10.1128/mbio.03062-23. Epub 2023 Dec 22. mBio. 2024. PMID: 38133430 Free PMC article.
-
Fungal Kinases With a Sweet Tooth: Pleiotropic Roles of Their Phosphorylated Inositol Sugar Products in the Pathogenicity of Cryptococcus neoformans Present Novel Drug Targeting Opportunities.Front Cell Infect Microbiol. 2019 Jul 15;9:248. doi: 10.3389/fcimb.2019.00248. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380293 Free PMC article. Review.
-
The inositol pyrophosphate metabolism of Dictyostelium discoideum does not regulate inorganic polyphosphate (polyP) synthesis.Adv Biol Regul. 2022 Jan;83:100835. doi: 10.1016/j.jbior.2021.100835. Epub 2021 Nov 10. Adv Biol Regul. 2022. PMID: 34782304 Free PMC article.
-
Genetic screen for suppression of transcriptional interference identifies a gain-of-function mutation in Pol2 termination factor Seb1.Proc Natl Acad Sci U S A. 2021 Aug 17;118(33):e2108105118. doi: 10.1073/pnas.2108105118. Proc Natl Acad Sci U S A. 2021. PMID: 34389684 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
