HAVCR1 (CD365) and Its Mouse Ortholog Are Functional Hepatitis A Virus (HAV) Cellular Receptors That Mediate HAV Infection
- PMID: 29437974
- PMCID: PMC5899186
- DOI: 10.1128/JVI.02065-17
HAVCR1 (CD365) and Its Mouse Ortholog Are Functional Hepatitis A Virus (HAV) Cellular Receptors That Mediate HAV Infection
Abstract
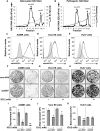
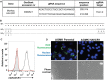
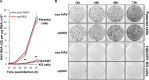
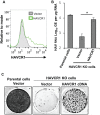
The hepatitis A virus (HAV) cellular receptor 1 (HAVCR1), classified as CD365, was initially discovered as an HAV cellular receptor using an expression cloning strategy. Due to the lack of HAV receptor-negative replication-competent cells, it was not possible to fully prove that HAVCR1 was a functional HAV receptor. However, biochemistry, classical virology, and epidemiology studies further supported the functional role of HAVCR1 as an HAV receptor. Here, we show that an anti-HAVCR1 monoclonal antibody that protected African green monkey kidney (AGMK) cells against HAV infection only partially protected monkey Vero E6 cells and human hepatoma Huh7 cells, indicating that these two cell lines express alternative yet unidentified HAV receptors. Therefore, we focused our work on AGMK cells to further characterize the function of HAVCR1 as an HAV receptor. Advances in clustered regularly interspaced short palindromic repeat/Cas9 technology allowed us to knock out the monkey ortholog of HAVCR1 in AGMK cells. The resulting AGMK HAVCR1 knockout (KO) cells lost susceptibility to HAV infection, including HAV-free viral particles (vpHAV) and exosomes purified from HAV-infected cells (exo-HAV). Transfection of HAVCR1 cDNA into AGMK HAVCR1 KO cells restored susceptibility to vpHAV and exo-HAV infection. Furthermore, transfection of the mouse ortholog of HAVCR1, mHavcr1, also restored the susceptibility of AGMK HAVCR1 KO cells to HAV infection. Taken together, our data clearly show that HAVCR1 and mHavcr1 are functional HAV receptors that mediate HAV infection. This work paves the way for the identification of alternative HAV receptors to gain a complete understanding of their interplay with HAVCR1 in the cell entry and pathogenic processes of HAV.IMPORTANCE HAVCR1, an HAV receptor, is expressed in different cell types, including regulatory immune cells and antigen-presenting cells. How HAV evades the immune response during a long incubation period of up to 4 weeks and the mechanism by which the subsequent necroinflammatory process clears the infection remain a puzzle that most likely involves the HAV-HAVCR1 interaction. Based on negative data, a recent paper from the S. M. Lemon and W. Maury laboratories (A. Das, A. Hirai-Yuki, O. Gonzalez-Lopez, B. Rhein, S. Moller-Tank, R. Brouillette, L. Hensley, I. Misumi, W. Lovell, J. M. Cullen, J. K. Whitmire, W. Maury, and S. M. Lemon, mBio 8:e00969-17, 2017, https://doi.org/10.1128/mBio.00969-17) suggested that HAVCR1 is not a functional HAV receptor, nor it is it required for HAV infection. However, our data, based on regain of the HAV receptor function in HAVCR1 knockout cells transfected with HAVCR1 cDNA, disagree with their findings. Our positive data show conclusively that HAVCR1 is indeed a functional HAV receptor and lays the ground for the identification of alternative HAV receptors and how they interact with HAVCR1 in cell entry and the pathogenesis of HAV.
Keywords: African green monkey cells; CD365; CRISPR/Cas9; HAVCR1; KIM1; TIM1; Vero E6 cells; cellular receptor; gene editing; hepatitis A virus; human hepatoma Huh7 cells; mouse ortholog.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures
Comment in
-
TIM1 (HAVCR1): an Essential "Receptor" or an "Accessory Attachment Factor" for Hepatitis A Virus?J Virol. 2019 May 15;93(11):e01793-18. doi: 10.1128/JVI.01793-18. Print 2019 Jun 1. J Virol. 2019. PMID: 31092684 Free PMC article. No abstract available.
-
Reply to Das et al., "TIM1 (HAVCR1): an Essential 'Receptor' or an 'Accessory Attachment Factor' for Hepatitis A Virus?".J Virol. 2019 May 15;93(11):e02040-18. doi: 10.1128/JVI.02040-18. Print 2019 Jun 1. J Virol. 2019. PMID: 31092685 Free PMC article. No abstract available.
Similar articles
-
TIM1 (HAVCR1) Is Not Essential for Cellular Entry of Either Quasi-enveloped or Naked Hepatitis A Virions.mBio. 2017 Sep 5;8(5):e00969-17. doi: 10.1128/mBio.00969-17. mBio. 2017. PMID: 28874468 Free PMC article.
-
Exosome mimicry by a HAVCR1-NPC1 pathway of endosomal fusion mediates hepatitis A virus infection.Nat Microbiol. 2020 Sep;5(9):1096-1106. doi: 10.1038/s41564-020-0740-y. Epub 2020 Jun 15. Nat Microbiol. 2020. PMID: 32541946 Free PMC article.
-
Determinants in the Ig Variable Domain of Human HAVCR1 (TIM-1) Are Required To Enhance Hepatitis C Virus Entry.J Virol. 2018 Feb 26;92(6):e01742-17. doi: 10.1128/JVI.01742-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29321304 Free PMC article.
-
Potential of CRISPR/Cas system as emerging tools in the detection of viral hepatitis infection.Virol J. 2023 May 8;20(1):91. doi: 10.1186/s12985-023-02048-5. Virol J. 2023. PMID: 37158910 Free PMC article. Review.
-
[Advances in application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 system in stem cells research].Zhonghua Shao Shang Za Zhi. 2018 Apr 20;34(4):253-256. doi: 10.3760/cma.j.issn.1009-2587.2018.04.013. Zhonghua Shao Shang Za Zhi. 2018. PMID: 29690746 Review. Chinese.
Cited by
-
CRISPR/Cas9 as a New Antiviral Strategy for Treating Hepatitis Viral Infections.Int J Mol Sci. 2023 Dec 26;25(1):334. doi: 10.3390/ijms25010334. Int J Mol Sci. 2023. PMID: 38203503 Free PMC article. Review.
-
The phosphatidylserine receptor TIM1 promotes infection of enveloped hepatitis E virus.Cell Mol Life Sci. 2023 Oct 13;80(11):326. doi: 10.1007/s00018-023-04977-4. Cell Mol Life Sci. 2023. PMID: 37833515 Free PMC article.
-
Hepatitis A: Viral Structure, Classification, Life Cycle, Clinical Symptoms, Diagnosis Error, and Vaccination.Can J Infect Dis Med Microbiol. 2023 Jan 4;2023:4263309. doi: 10.1155/2023/4263309. eCollection 2023. Can J Infect Dis Med Microbiol. 2023. PMID: 36644336 Free PMC article. Review.
-
Pathogenicity and virulence of hepatitis A virus.Virulence. 2021 Dec;12(1):1174-1185. doi: 10.1080/21505594.2021.1910442. Virulence. 2021. PMID: 33843464 Free PMC article. Review.
-
Recent Progress on Exosomes in RNA Virus Infection.Viruses. 2021 Feb 8;13(2):256. doi: 10.3390/v13020256. Viruses. 2021. PMID: 33567490 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

