Prefoldin, a jellyfish-like molecular chaperone: functional cooperation with a group II chaperonin and beyond
- PMID: 29427249
- PMCID: PMC5899742
- DOI: 10.1007/s12551-018-0400-0
Prefoldin, a jellyfish-like molecular chaperone: functional cooperation with a group II chaperonin and beyond
Abstract
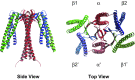
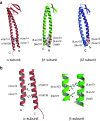
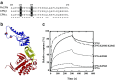
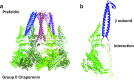
Prefoldin is a hexameric molecular chaperone found in the cytosol of archaea and eukaryotes. Its hexameric complex is built from two related classes of subunits and has the appearance of a jellyfish: its body consists of a double beta-barrel assembly with six long tentacle-like coiled coils protruding from it. Using the tentacles, prefoldin captures an unfolded protein substrate and transfers it to a group II chaperonin. The prefoldin-group II chaperonin system is thought to be important for the folding of newly synthesized proteins and for their maintenance, or proteostasis, in the cytosol. Based on structural information of archaeal prefoldins, the mechanisms of substrate recognition and prefoldin-chaperonin cooperation have been investigated. In contrast, the role and mechanism of eukaryotic PFDs remain unknown. Recent studies have shown that prefoldin plays an important role in proteostasis and is involved in various diseases. In this paper, we review a series of studies on the molecular mechanisms of archaeal prefoldins and introduce recent findings about eukaryotic prefoldin.
Keywords: Chaperonin; Molecular chaperone; Prefoldin.
Conflict of interest statement
Conflict of interest
Muhamad Sahlan declares that he has no conflict of interest. Tamotsu Zako declares that he has no conflict of interest. Masafumi Yohda declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
Similar articles
-
Expression, Functional Characterization, and Preliminary Crystallization of the Cochaperone Prefoldin from the Thermophilic Fungus Chaetomium thermophilum.Int J Mol Sci. 2018 Aug 19;19(8):2452. doi: 10.3390/ijms19082452. Int J Mol Sci. 2018. PMID: 30126249 Free PMC article.
-
Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins.Cell. 2000 Nov 10;103(4):621-32. doi: 10.1016/s0092-8674(00)00165-3. Cell. 2000. PMID: 11106732
-
Analysis of the interaction mode between hyperthermophilic archaeal group II chaperonin and prefoldin using a platform of chaperonin oligomers of various subunit arrangements.Biochim Biophys Acta. 2010 Sep;1804(9):1810-6. doi: 10.1016/j.bbapap.2010.04.013. Epub 2010 May 6. Biochim Biophys Acta. 2010. PMID: 20451672
-
Structure and function of archaeal prefoldin, a co-chaperone of group II chaperonin.Front Biosci (Landmark Ed). 2010 Jan 1;15(2):708-17. doi: 10.2741/3641. Front Biosci (Landmark Ed). 2010. PMID: 20036841 Review.
-
Prefoldins in Archaea.Adv Exp Med Biol. 2018;1106:11-23. doi: 10.1007/978-3-030-00737-9_2. Adv Exp Med Biol. 2018. PMID: 30484150 Review.
Cited by
-
Defining heat shock response for the thermoacidophilic model crenarchaeon Sulfolobus acidocaldarius.Extremophiles. 2020 Sep;24(5):681-692. doi: 10.1007/s00792-020-01184-y. Epub 2020 Jun 19. Extremophiles. 2020. PMID: 32562000
-
Foreword to 'Multiscale structural biology: biophysical principles and mechanisms underlying the action of bio-nanomachines', a special issue in Honour of Fumio Arisaka's 70th birthday.Biophys Rev. 2018 Apr;10(2):105-129. doi: 10.1007/s12551-018-0401-z. Epub 2018 Mar 2. Biophys Rev. 2018. PMID: 29500796 Free PMC article.
-
Ligand Binding Site Structure Shapes Folding, Assembly and Degradation of Homomeric Protein Complexes.J Mol Biol. 2019 Sep 6;431(19):3871-3888. doi: 10.1016/j.jmb.2019.07.014. Epub 2019 Jul 12. J Mol Biol. 2019. PMID: 31306664 Free PMC article.
-
Prefoldins contribute to maintaining the levels of the spliceosome LSM2-8 complex through Hsp90 in Arabidopsis.Nucleic Acids Res. 2020 Jun 19;48(11):6280-6293. doi: 10.1093/nar/gkaa354. Nucleic Acids Res. 2020. PMID: 32396196 Free PMC article.
-
PFDN4 as a Prognostic Marker Was Associated with Chemotherapy Resistance through CREBP1/AURKA Pathway in Triple-Negative Breast Cancer.Int J Mol Sci. 2024 Mar 31;25(7):3906. doi: 10.3390/ijms25073906. Int J Mol Sci. 2024. PMID: 38612711 Free PMC article.
References
-
- Fujiwara S, Aki R, Yoshida M, Higashibata H, Imanaka T, Fukuda W. Expression profiles and physiological roles of two types of molecular chaperonins from the hyperthermophilic archaeon Thermococcus kodakarensis. Appl Environ Microbiol. 2008;74:7306–7312. doi: 10.1128/AEM.01245-08. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

