Post-translational regulation of the maternal-to-zygotic transition
- PMID: 29427077
- PMCID: PMC11105290
- DOI: 10.1007/s00018-018-2750-y
Post-translational regulation of the maternal-to-zygotic transition
Abstract
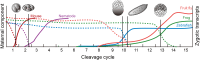
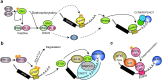
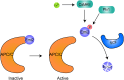
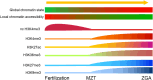
The maternal-to-zygotic transition (MZT) is essential for the developmental control handed from maternal products to newly synthesized zygotic genome in the earliest stages of embryogenesis, including maternal component (mRNAs and proteins) degradation and zygotic genome activation (ZGA). Various protein post-translational modifications have been identified during the MZT, such as phosphorylation, methylation and ubiquitination. Precise post-translational regulation mechanisms are essential for the timely transition of early embryonic development. In this review, we summarize recent progress regarding the molecular mechanisms underlying post-translational regulation of maternal component degradation and ZGA during the MZT and discuss some important issues in the field.
Keywords: Histone modification; MZT; Phosphorylation; Ubiquitination; ZGA.
Figures
Similar articles
-
The maternal-to-zygotic transition revisited.Development. 2019 Jun 12;146(11):dev161471. doi: 10.1242/dev.161471. Development. 2019. PMID: 31189646 Review.
-
A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals†.Biol Reprod. 2019 Sep 1;101(3):579-590. doi: 10.1093/biolre/ioz012. Biol Reprod. 2019. PMID: 30715134 Review.
-
Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition.Sci Adv. 2022 Feb 4;8(5):eabj3967. doi: 10.1126/sciadv.abj3967. Epub 2022 Feb 2. Sci Adv. 2022. PMID: 35108058 Free PMC article.
-
The maternal-to-zygotic transition.Curr Biol. 2024 Jun 3;34(11):R519-R523. doi: 10.1016/j.cub.2024.04.044. Curr Biol. 2024. PMID: 38834020 Review.
-
Developmental series of gene expression clarifies maternal mRNA provisioning and maternal-to-zygotic transition in a reef-building coral.BMC Genomics. 2021 Nov 11;22(1):815. doi: 10.1186/s12864-021-08114-y. BMC Genomics. 2021. PMID: 34763678 Free PMC article.
Cited by
-
The neglected part of early embryonic development: maternal protein degradation.Cell Mol Life Sci. 2020 Aug;77(16):3177-3194. doi: 10.1007/s00018-020-03482-2. Epub 2020 Feb 24. Cell Mol Life Sci. 2020. PMID: 32095869 Free PMC article. Review.
-
Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens.Int J Mol Sci. 2020 Oct 31;21(21):8170. doi: 10.3390/ijms21218170. Int J Mol Sci. 2020. PMID: 33142918 Free PMC article.
-
Zygotic genome activation in the chicken: a comparative review.Cell Mol Life Sci. 2020 May;77(10):1879-1891. doi: 10.1007/s00018-019-03360-6. Epub 2019 Nov 15. Cell Mol Life Sci. 2020. PMID: 31728579 Free PMC article. Review.
-
ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation.Nucleic Acids Res. 2019 Dec 2;47(21):11387-11402. doi: 10.1093/nar/gkz863. Nucleic Acids Res. 2019. PMID: 31598710 Free PMC article.
-
Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans.Nat Commun. 2020 Oct 1;11(1):4917. doi: 10.1038/s41467-020-18680-6. Nat Commun. 2020. PMID: 33004802 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 2016YFA0500901/National Key R&D Program of China/International
- 31471277/National Natural Science Foundation of China/International
- 91649202/National Natural Science Foundation of China/International
- 31471107/National Natural Science Foundation of China/International
- 31771501/National Natural Science Foundation of China/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

