Rce1: mechanism and inhibition
- PMID: 29424242
- PMCID: PMC5874806
- DOI: 10.1080/10409238.2018.1431606
Rce1: mechanism and inhibition
Abstract
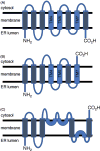
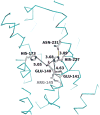
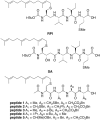
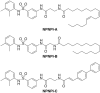
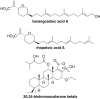
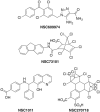
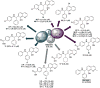
Ras converting enzyme 1 (Rce1) is an integral membrane endoprotease localized to the endoplasmic reticulum that mediates the cleavage of the carboxyl-terminal three amino acids from CaaX proteins, whose members play important roles in cell signaling processes. Examples include the Ras family of small GTPases, the γ-subunit of heterotrimeric GTPases, nuclear lamins, and protein kinases and phosphatases. CaaX proteins, especially Ras, have been implicated in cancer, and understanding the post-translational modifications of CaaX proteins would provide insight into their biological function and regulation. Many proteolytic mechanisms have been proposed for Rce1, but sequence alignment, mutational studies, topology, and recent crystallographic data point to a novel mechanism involving a glutamate-activated water and an oxyanion hole. Studies using in vivo and in vitro reporters of Rce1 activity have revealed that the enzyme cleaves only prenylated substrates and the identity of the a2 amino residue in the Ca1a2X sequence is most critical for recognition, preferring Ile, Leu, or Val. Substrate mimetics can be somewhat effective inhibitors of Rce1 in vitro. Small-molecule inhibitor discovery is currently limited by the lack of structural information on a eukaryotic enzyme, but a set of 8-hydroxyquinoline derivatives has demonstrated an ability to mislocalize all three mammalian Ras isoforms, giving optimism that potent, selective inhibitors might be developed. Much remains to be discovered regarding cleavage specificity, the impact of chemical inhibition, and the potential of Rce1 as a therapeutic target, not only for cancer, but also for other diseases.
Keywords: CaaX proteins; Ras; Ras converting enzyme; cancer; proteases.
Conflict of interest statement
The authors report no conflicts of interest.
Figures

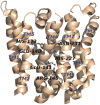
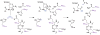

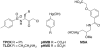
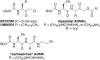
Similar articles
-
Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1.Nature. 2013 Dec 12;504(7479):301-5. doi: 10.1038/nature12754. Epub 2013 Dec 1. Nature. 2013. PMID: 24291792 Free PMC article.
-
8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells.Bioorg Med Chem. 2016 Jan 15;24(2):160-78. doi: 10.1016/j.bmc.2015.11.043. Epub 2015 Nov 30. Bioorg Med Chem. 2016. PMID: 26706114 Free PMC article.
-
Targeted genetic and small molecule disruption of N-Ras CaaX cleavage alters its localization and oncogenic potential.Bioorg Chem. 2024 Jun;147:107316. doi: 10.1016/j.bioorg.2024.107316. Epub 2024 Mar 27. Bioorg Chem. 2024. PMID: 38583246
-
Genetic analyses of the role of RCE1 in RAS membrane association and transformation.Methods Enzymol. 2008;438:367-89. doi: 10.1016/S0076-6879(07)38026-9. Methods Enzymol. 2008. PMID: 18413262 Free PMC article. Review.
-
CaaX converting enzymes.Curr Opin Lipidol. 1998 Apr;9(2):99-102. doi: 10.1097/00041433-199804000-00004. Curr Opin Lipidol. 1998. PMID: 9559265 Review.
Cited by
-
Protein Isoprenylation in Yeast Targets COOH-Terminal Sequences Not Adhering to the CaaX Consensus.Genetics. 2018 Dec;210(4):1301-1316. doi: 10.1534/genetics.118.301454. Epub 2018 Sep 26. Genetics. 2018. PMID: 30257935 Free PMC article.
-
A Novel KRAS Antibody Highlights a Regulation Mechanism of Post-Translational Modifications of KRAS during Tumorigenesis.Int J Mol Sci. 2020 Sep 2;21(17):6361. doi: 10.3390/ijms21176361. Int J Mol Sci. 2020. PMID: 32887255 Free PMC article.
-
RAS-targeted therapies: is the undruggable drugged?Nat Rev Drug Discov. 2020 Aug;19(8):533-552. doi: 10.1038/s41573-020-0068-6. Epub 2020 Jun 11. Nat Rev Drug Discov. 2020. PMID: 32528145 Free PMC article. Review.
-
Biology, pathology, and therapeutic targeting of RAS.Adv Cancer Res. 2020;148:69-146. doi: 10.1016/bs.acr.2020.05.002. Epub 2020 Jul 9. Adv Cancer Res. 2020. PMID: 32723567 Free PMC article. Review.
-
Protein lipidation in health and disease: molecular basis, physiological function and pathological implication.Signal Transduct Target Ther. 2024 Mar 15;9(1):60. doi: 10.1038/s41392-024-01759-7. Signal Transduct Target Ther. 2024. PMID: 38485938 Free PMC article. Review.
References
-
- Baell JB. Feeling nature’s PAINS: natural products, natural product drugs, and pan assay interference compounds (PAINS) J Nat Prod. 2016;79:616–628. - PubMed
-
- Baell JB, Ferrins L, Falk H, Nikolakopoulos G. PAINS: relevance to tool compound discovery and fragment-based screening. Aust J Chem. 2013;66:1483–1494.
-
- Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. - PubMed
-
- Baell JB, Walters MA. Chemistry: chemical con artists foil drug discovery. Nature. 2014;513:481–483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
