The central role of DNA damage and repair in CAG repeat diseases
- PMID: 29419417
- PMCID: PMC5818082
- DOI: 10.1242/dmm.031930
The central role of DNA damage and repair in CAG repeat diseases
Abstract
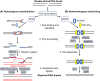
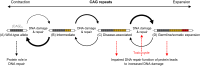
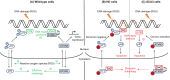
Diseases such as Huntington's disease and certain spinocerebellar ataxias are caused by the expansion of genomic cytosine-adenine-guanine (CAG) trinucleotide repeats beyond a specific threshold. These diseases are all characterised by neurological symptoms and central neurodegeneration, but our understanding of how expanded repeats drive neuronal loss is incomplete. Recent human genetic evidence implicates DNA repair pathways, especially mismatch repair, in modifying the onset and progression of CAG repeat diseases. Repair pathways might operate directly on repeat sequences by licensing or inhibiting repeat expansion in neurons. Alternatively, or in addition, because many of the genes containing pathogenic CAG repeats encode proteins that themselves have roles in the DNA damage response, it is possible that repeat expansions impair specific DNA repair pathways. DNA damage could then accrue in neurons, leading to further expansion at repeat loci, thus setting up a vicious cycle of pathology. In this review, we consider DNA damage and repair pathways in postmitotic neurons in the context of disease-causing CAG repeats. Investigating and understanding these pathways, which are clearly relevant in promoting and ameliorating disease in humans, is a research priority, as they are known to modify disease and therefore constitute prevalidated drug targets.
Keywords: CAG repeat; DNA damage; DNA repair; Huntington's disease; Spinocerebellar ataxia.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

Similar articles
-
DNA repair in the trinucleotide repeat disorders.Lancet Neurol. 2017 Jan;16(1):88-96. doi: 10.1016/S1474-4422(16)30350-7. Lancet Neurol. 2017. PMID: 27979358 Review.
-
DNA repair in trinucleotide repeat ataxias.FEBS J. 2018 Oct;285(19):3669-3682. doi: 10.1111/febs.14644. Epub 2018 Sep 10. FEBS J. 2018. PMID: 30152109 Review.
-
What is the Pathogenic CAG Expansion Length in Huntington's Disease?J Huntingtons Dis. 2021;10(1):175-202. doi: 10.3233/JHD-200445. J Huntingtons Dis. 2021. PMID: 33579866 Free PMC article. Review.
-
Methods for Assessing DNA Repair and Repeat Expansion in Huntington's Disease.Methods Mol Biol. 2018;1780:483-495. doi: 10.1007/978-1-4939-7825-0_22. Methods Mol Biol. 2018. PMID: 29856032 Review.
-
Expanded CAG/CTG repeat DNA induces a checkpoint response that impacts cell proliferation in Saccharomyces cerevisiae.PLoS Genet. 2011 Mar;7(3):e1001339. doi: 10.1371/journal.pgen.1001339. Epub 2011 Mar 17. PLoS Genet. 2011. PMID: 21437275 Free PMC article.
Cited by
-
Age-related and disease locus-specific mechanisms contribute to early remodelling of chromatin structure in Huntington's disease mice.Nat Commun. 2021 Jan 13;12(1):364. doi: 10.1038/s41467-020-20605-2. Nat Commun. 2021. PMID: 33441541 Free PMC article.
-
DNA Repair in Huntington's Disease and Spinocerebellar Ataxias: Somatic Instability and Alternative Hypotheses.J Huntingtons Dis. 2021;10(1):165-173. doi: 10.3233/JHD-200414. J Huntingtons Dis. 2021. PMID: 33579859 Free PMC article. Review.
-
HSF1 and Its Role in Huntington's Disease Pathology.Adv Exp Med Biol. 2023;1410:35-95. doi: 10.1007/5584_2022_742. Adv Exp Med Biol. 2023. PMID: 36396925 Review.
-
Keeping ribosomal DNA intact: a repeating challenge.Chromosome Res. 2019 Mar;27(1-2):57-72. doi: 10.1007/s10577-018-9594-z. Epub 2018 Dec 17. Chromosome Res. 2019. PMID: 30556094 Free PMC article. Review.
-
MSH7 confers quantitative variation in pollen fertility and boosts grain yield in maize.Plant Biotechnol J. 2024 May;22(5):1372-1386. doi: 10.1111/pbi.14272. Epub 2024 Jan 23. Plant Biotechnol J. 2024. PMID: 38263872 Free PMC article.
References
-
- Abraham K. J., Chan J. N. Y., Salvi J. S., Ho B., Hall A., Vidya E., Guo R., Killackey S. A., Liu N., Lee J. E. et al. (2016). Intersection of calorie restriction and magnesium in the suppression of genome-destabilizing RNA–DNA hybrids. Nucleic Acids Res. 44, 8870-8884. 10.1093/nar/gkw752 - DOI - PMC - PubMed
-
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21 10.1186/s13229-017-0137-9 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

