miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma
- PMID: 29416013
- PMCID: PMC5833358
- DOI: 10.1038/s41419-017-0258-2
miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma
Abstract
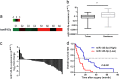
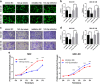
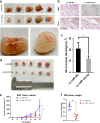
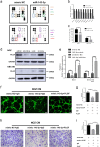
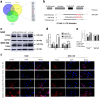
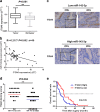
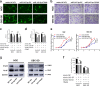
Gallbladder cancer (GBC) is the most common malignant tumour of the biliary track system. Angiogenesis plays a pivotal role in the development and progression of malignant tumours. miR-143-3p acts as a tumour suppressor in various cancers. Their role in GBC is however less well defined. Here we show that the expression levels of miR-143-3p were decreased in human GBC tissues compared with the non-tumour adjacent tissue (NAT) counterparts and were closely associated with overall survival. We discovered that miR-143-3p was a novel inhibitor of tumour growth and angiogenesis in vivo and in vitro. Our antibody array, ELISA and PLGF rescue analyses indicated that PLGF played an essential role in the antiangiogenic effect of miR-143-3p. Furthermore, we used miRNA target-prediction software and dual-luciferase assays to confirm that integrin α6 (ITGA6) acted as a direct target of miR-143-3p. Our ELISA and western blot analyses confirmed that the expression of PLGF was decreased via the ITGA6/PI3K/AKT pathway. In conclusion, miR-143-3p suppresses tumour angiogenesis and growth of GBC through the ITGA6/PI3K/AKT/PLGF pathways and may be a novel molecular therapeutic target for GBC.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Overexpressed microRNA-136 works as a cancer suppressor in gallbladder cancer through suppression of JNK signaling pathway via inhibition of MAP2K4.Am J Physiol Gastrointest Liver Physiol. 2019 Nov 1;317(5):G670-G681. doi: 10.1152/ajpgi.00055.2019. Epub 2019 Aug 1. Am J Physiol Gastrointest Liver Physiol. 2019. PMID: 31369289
-
LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis.Mol Cancer. 2019 Nov 21;18(1):167. doi: 10.1186/s12943-019-1097-9. Mol Cancer. 2019. PMID: 31752906 Free PMC article.
-
MiR-6875-3p promotes the proliferation, invasion and metastasis of hepatocellular carcinoma via BTG2/FAK/Akt pathway.J Exp Clin Cancer Res. 2019 Jan 8;38(1):7. doi: 10.1186/s13046-018-1020-z. J Exp Clin Cancer Res. 2019. PMID: 30621734 Free PMC article.
-
Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling.J Exp Clin Cancer Res. 2018 Jul 18;37(1):160. doi: 10.1186/s13046-018-0813-4. J Exp Clin Cancer Res. 2018. PMID: 30021600 Free PMC article.
-
The role of PI3K/AKT signaling pathway in gallbladder carcinoma.Am J Transl Res. 2022 Jul 15;14(7):4426-4442. eCollection 2022. Am J Transl Res. 2022. PMID: 35958463 Free PMC article. Review.
Cited by
-
Biological functions and potential mechanisms of miR‑143‑3p in cancers (Review).Oncol Rep. 2024 Sep;52(3):113. doi: 10.3892/or.2024.8772. Epub 2024 Jul 12. Oncol Rep. 2024. PMID: 38994765 Free PMC article. Review.
-
Discovery and Functional Analysis of Secondary Hair Follicle miRNAs during Annual Cashmere Growth.Int J Mol Sci. 2023 Jan 5;24(2):1063. doi: 10.3390/ijms24021063. Int J Mol Sci. 2023. PMID: 36674578 Free PMC article.
-
Footprints of microRNAs in Cancer Biology.Biomedicines. 2021 Oct 19;9(10):1494. doi: 10.3390/biomedicines9101494. Biomedicines. 2021. PMID: 34680611 Free PMC article. Review.
-
High expression of PSMC2 promotes gallbladder cancer through regulation of GNG4 and predicts poor prognosis.Oncogenesis. 2021 May 20;10(5):43. doi: 10.1038/s41389-021-00330-1. Oncogenesis. 2021. PMID: 34016944 Free PMC article.
-
Long non-coding RNA Taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF-1α via miR-143-5p.Cell Death Dis. 2019 Mar 25;10(4):280. doi: 10.1038/s41419-019-1509-1. Cell Death Dis. 2019. PMID: 30911001 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

