Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome
- PMID: 29410130
- PMCID: PMC6023675
- DOI: 10.1016/j.jmb.2018.01.016
Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome
Abstract
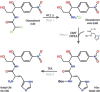
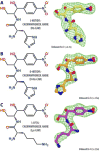
Antibiotic chloramphenicol (CHL) binds with a moderate affinity at the peptidyl transferase center of the bacterial ribosome and inhibits peptide bond formation. As an approach for modifying and potentially improving properties of this inhibitor, we explored ribosome binding and inhibitory activity of a number of amino acid analogs of CHL. The L-histidyl analog binds to the ribosome with the affinity exceeding that of CHL by 10 fold. Several of the newly synthesized analogs were able to inhibit protein synthesis and exhibited the mode of action that was distinct from the action of CHL. However, the inhibitory properties of the semi-synthetic CHL analogs did not correlate with their affinity and in general, the amino acid analogs of CHL were less active inhibitors of translation in comparison with the original antibiotic. The X-ray crystal structures of the Thermus thermophilus 70S ribosome in complex with three semi-synthetic analogs showed that CHL derivatives bind at the peptidyl transferase center, where the aminoacyl moiety of the tested compounds established idiosyncratic interactions with rRNA. Although still fairly inefficient inhibitors of translation, the synthesized compounds represent promising chemical scaffolds that target the peptidyl transferase center of the ribosome and potentially are suitable for further exploration.
Keywords: X-ray structure; antibiotic; peptidyl transferase center; protein synthesis; ribosome.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
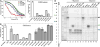
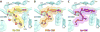
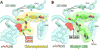
Similar articles
-
High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.RNA. 2019 May;25(5):600-606. doi: 10.1261/rna.069260.118. Epub 2019 Feb 7. RNA. 2019. PMID: 30733327 Free PMC article.
-
Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.Structure. 2024 Sep 5;32(9):1429-1442.e6. doi: 10.1016/j.str.2024.06.013. Epub 2024 Jul 16. Structure. 2024. PMID: 39019034
-
Binding and Action of Triphenylphosphonium Analog of Chloramphenicol upon the Bacterial Ribosome.Antibiotics (Basel). 2021 Apr 5;10(4):390. doi: 10.3390/antibiotics10040390. Antibiotics (Basel). 2021. PMID: 33916420 Free PMC article.
-
On the specificity of antibiotics targeting the large ribosomal subunit.Ann N Y Acad Sci. 2011 Dec;1241:1-16. doi: 10.1111/j.1749-6632.2011.06192.x. Ann N Y Acad Sci. 2011. PMID: 22191523 Review.
-
Insights into protein biosynthesis and ribosome function through inhibitors.Prog Nucleic Acid Res Mol Biol. 1976;17:217-45. doi: 10.1016/s0079-6603(08)60071-9. Prog Nucleic Acid Res Mol Biol. 1976. PMID: 778922 Review. No abstract available.
Cited by
-
Hydrocinnamic Acid and Perillyl Alcohol Potentiate the Action of Antibiotics against Escherichia coli.Antibiotics (Basel). 2023 Feb 9;12(2):360. doi: 10.3390/antibiotics12020360. Antibiotics (Basel). 2023. PMID: 36830271 Free PMC article.
-
New Chloramphenicol Derivatives with a Modified Dichloroacetyl Tail as Potential Antimicrobial Agents.Antibiotics (Basel). 2021 Apr 6;10(4):394. doi: 10.3390/antibiotics10040394. Antibiotics (Basel). 2021. PMID: 33917453 Free PMC article.
-
Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.Nat Chem Biol. 2024 Jul;20(7):867-876. doi: 10.1038/s41589-023-01525-w. Epub 2024 Jan 18. Nat Chem Biol. 2024. PMID: 38238495
-
New Chloramphenicol Derivatives from the Viewpoint of Anticancer and Antimicrobial Activity.Antibiotics (Basel). 2019 Jan 29;8(1):9. doi: 10.3390/antibiotics8010009. Antibiotics (Basel). 2019. PMID: 30699905 Free PMC article.
-
Antibiotics targeting bacterial ribosomal subunit biogenesis.J Antimicrob Chemother. 2020 Apr 1;75(4):787-806. doi: 10.1093/jac/dkz544. J Antimicrob Chemother. 2020. PMID: 31942624 Free PMC article. Review.
References
-
- Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol. 2005;40:285–311. - PubMed
-
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. - PubMed
-
- McKie SA. Antibiotics: where to throw the spanner in the ribosomal machinery? Future Med Chem. 2016;8:1981–2002. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

