Structure of the Deactive State of Mammalian Respiratory Complex I
- PMID: 29395787
- PMCID: PMC5807054
- DOI: 10.1016/j.str.2017.12.014
Structure of the Deactive State of Mammalian Respiratory Complex I
Abstract
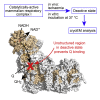
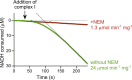
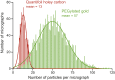
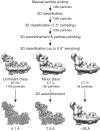
Complex I (NADH:ubiquinone oxidoreductase) is central to energy metabolism in mammalian mitochondria. It couples NADH oxidation by ubiquinone to proton transport across the energy-conserving inner membrane, catalyzing respiration and driving ATP synthesis. In the absence of substrates, active complex I gradually enters a pronounced resting or deactive state. The active-deactive transition occurs during ischemia and is crucial for controlling how respiration recovers upon reperfusion. Here, we set a highly active preparation of Bos taurus complex I into the biochemically defined deactive state, and used single-particle electron cryomicroscopy to determine its structure to 4.1 Å resolution. We show that the deactive state arises when critical structural elements that form the ubiquinone-binding site become disordered, and we propose reactivation is induced when substrate binding to the NADH-reduced enzyme templates their reordering. Our structure both rationalizes biochemical data on the deactive state and offers new insights into its physiological and cellular roles.
Keywords: NADH:ubiquinone oxidoreductase; PEGylated gold grid; cryo-EM; disordered protein structure; electron transport chain; membrane protein; mitochondria.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
The deactive form of respiratory complex I from mammalian mitochondria is a Na+/H+ antiporter.J Biol Chem. 2012 Oct 5;287(41):34743-51. doi: 10.1074/jbc.M112.384560. Epub 2012 Aug 1. J Biol Chem. 2012. PMID: 22854968 Free PMC article.
-
Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.Elife. 2023 Jan 9;12:e84424. doi: 10.7554/eLife.84424. Elife. 2023. PMID: 36622099 Free PMC article.
-
Molecular strain in the active/deactive-transition modulates domain coupling in respiratory complex I.Biochim Biophys Acta Bioenerg. 2021 May 1;1862(5):148382. doi: 10.1016/j.bbabio.2021.148382. Epub 2021 Jan 26. Biochim Biophys Acta Bioenerg. 2021. PMID: 33513365
-
Mammalian Respiratory Complex I Through the Lens of Cryo-EM.Annu Rev Biophys. 2019 May 6;48:165-184. doi: 10.1146/annurev-biophys-052118-115704. Epub 2019 Feb 20. Annu Rev Biophys. 2019. PMID: 30786232 Review.
-
Challenges in elucidating structure and mechanism of proton pumping NADH:ubiquinone oxidoreductase (complex I).J Bioenerg Biomembr. 2008 Oct;40(5):475-83. doi: 10.1007/s10863-008-9171-9. Epub 2008 Nov 4. J Bioenerg Biomembr. 2008. PMID: 18982432 Review.
Cited by
-
Structure of the turnover-ready state of an ancestral respiratory complex I.Nat Commun. 2024 Oct 29;15(1):9340. doi: 10.1038/s41467-024-53679-3. Nat Commun. 2024. PMID: 39472559 Free PMC article.
-
Mechanisms of Mitochondrial ROS Production in Assisted Reproduction: The Known, the Unknown, and the Intriguing.Antioxidants (Basel). 2020 Sep 29;9(10):933. doi: 10.3390/antiox9100933. Antioxidants (Basel). 2020. PMID: 33003362 Free PMC article. Review.
-
Interaction of human erythrocyte catalase with air-water interface in cryoEM.Microscopy (Oxf). 2022 Feb 18;71(Supplement_1):i51-i59. doi: 10.1093/jmicro/dfab037. Microscopy (Oxf). 2022. PMID: 35275189 Free PMC article.
-
Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.Science. 2023 Jan 27;379(6630):351-357. doi: 10.1126/science.ade3332. Epub 2023 Jan 26. Science. 2023. PMID: 36701435 Free PMC article.
-
Analysis of the assembly pathway for membrane subunits of Complex I reveals that subunit L (ND5) can assemble last in E. coli.BBA Adv. 2021;1:100027. doi: 10.1016/j.bbadva.2021.100027. Epub 2021 Oct 17. BBA Adv. 2021. PMID: 35814529 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

