Interferon regulatory factor 8 regulates caspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction
- PMID: 29357389
- PMCID: PMC5794192
- DOI: 10.1371/journal.ppat.1006868
Interferon regulatory factor 8 regulates caspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction
Abstract
Interferon regulatory factor 8 (IRF8), also known as interferon consensus sequence-binding protein (ICSBP), is a transcription factor of the IRF family. IRF8 plays a key role in normal B cell differentiation, a cellular process that is intrinsically associated with Epstein-Barr virus (EBV) reactivation. However, whether IRF8 regulates EBV lytic replication remains unknown. In this study, we utilized a CRISPR/Cas9 genomic editing approach to deplete IRF8 and found that IRF8 depletion dramatically inhibits the reactivation of EBV upon lytic induction. We demonstrated that IRF8 depletion suppresses the expression of a group of genes involved in apoptosis and thus inhibits apoptosis induction upon lytic induction by B cell receptor (BCR) stimulation or chemical induction. The protein levels of caspase-1, caspase-3 and caspase-8 all dramatically decreased in IRF8-depleted cells, which led to reduced caspase activation and the stabilization of KAP1, PAX5 and DNMT3A upon BCR stimulation. Interestingly, caspase inhibition blocked the degradation of KAP1, PAX5 and DNMT3A, suppressed EBV lytic gene expression and viral DNA replication upon lytic induction, suggesting that the reduced caspase expression in IRF8-depleted cells contributes to the suppression of EBV lytic replication. We further demonstrated that IRF8 directly regulates CASP1 (caspase-1) gene expression through targeting its gene promoter and knockdown of caspase-1 abrogates EBV reactivation upon lytic induction, partially through the stabilization of KAP1. Together our study suggested that, by modulating the activation of caspases and the subsequent cleavage of KAP1 upon lytic induction, IRF8 plays a critical role in EBV lytic reactivation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



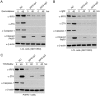
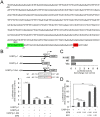
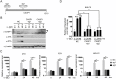

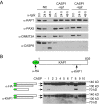

Similar articles
-
Protein inhibitor of activated STAT1 (PIAS1) inhibits IRF8 activation of Epstein-Barr virus lytic gene expression.Virology. 2020 Jan 15;540:75-87. doi: 10.1016/j.virol.2019.11.011. Epub 2019 Nov 11. Virology. 2020. PMID: 31743858 Free PMC article.
-
IRF4 promotes Epstein-Barr virus activation in Burkitt's lymphoma cells.J Gen Virol. 2019 May;100(5):851-862. doi: 10.1099/jgv.0.001249. Epub 2019 Mar 25. J Gen Virol. 2019. PMID: 30907723
-
Interferon-γ-inducible protein 16 (IFI16) is required for the maintenance of Epstein-Barr virus latency.Virol J. 2017 Nov 13;14(1):221. doi: 10.1186/s12985-017-0891-5. Virol J. 2017. PMID: 29132393 Free PMC article.
-
Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases.Autoimmunity. 2008 May;41(4):298-328. doi: 10.1080/08916930802024772. Autoimmunity. 2008. PMID: 18432410 Review.
-
Epstein-Barr Virus Lytic Cycle Reactivation.Curr Top Microbiol Immunol. 2015;391:237-61. doi: 10.1007/978-3-319-22834-1_8. Curr Top Microbiol Immunol. 2015. PMID: 26428377 Review.
Cited by
-
MYC Controls the Epstein-Barr Virus Lytic Switch.Mol Cell. 2020 May 21;78(4):653-669.e8. doi: 10.1016/j.molcel.2020.03.025. Epub 2020 Apr 20. Mol Cell. 2020. PMID: 32315601 Free PMC article.
-
Recurrent tonsillitis and parental perceptions of tonsillectomy during the COVID-19 pandemic.Int J Pediatr Otorhinolaryngol. 2020 Dec;139:110463. doi: 10.1016/j.ijporl.2020.110463. Epub 2020 Oct 23. Int J Pediatr Otorhinolaryngol. 2020. PMID: 33120105 Free PMC article.
-
Transcriptional Regulation of Inflammasomes.Int J Mol Sci. 2020 Oct 29;21(21):8087. doi: 10.3390/ijms21218087. Int J Mol Sci. 2020. PMID: 33138274 Free PMC article. Review.
-
Using proteolysis-targeting chimera technology to reduce navitoclax platelet toxicity and improve its senolytic activity.Nat Commun. 2020 Apr 24;11(1):1996. doi: 10.1038/s41467-020-15838-0. Nat Commun. 2020. PMID: 32332723 Free PMC article.
-
PIAS1 potentiates the anti-EBV activity of SAMHD1 through SUMOylation.Cell Biosci. 2021 Jul 8;11(1):127. doi: 10.1186/s13578-021-00636-y. Cell Biosci. 2021. PMID: 34238351 Free PMC article.
References
-
- Young LS, Yap LF, Murray PG (2016) Epstein-Barr virus: more than 50 years old and still providing surprises. Nature Reviews Cancer 16: 789–802. doi: 10.1038/nrc.2016.92 - DOI - PubMed
-
- Hammerschmidt W, Sugden B (2013) Replication of Epstein–Barr Viral DNA. Cold Spring Harbor perspectives in biology 5: a013029 doi: 10.1101/cshperspect.a013029 - DOI - PMC - PubMed
-
- Wu FY, Wang SE, Chen H, Wang L, Hayward SD, et al. (2004) CCAAT/enhancer binding protein alpha binds to the Epstein-Barr virus (EBV) ZTA protein through oligomeric interactions and contributes to cooperative transcriptional activation of the ZTA promoter through direct binding to the ZII and ZIIIB motifs during induction of the EBV lytic cycle. J Virol 78: 4847–4865. doi: 10.1128/JVI.78.9.4847-4865.2004 - DOI - PMC - PubMed
-
- Robinson AR, Kwek SS, Kenney SC (2012) The B-cell specific transcription factor, Oct-2, promotes Epstein-Barr virus latency by inhibiting the viral immediate-early protein, BZLF1. PLoS Pathog 8: e1002516 doi: 10.1371/journal.ppat.1002516 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

