Interactions of 2'-O-methyl oligoribonucleotides with the RNA models of the 30S subunit A-site
- PMID: 29351348
- PMCID: PMC5774723
- DOI: 10.1371/journal.pone.0191138
Interactions of 2'-O-methyl oligoribonucleotides with the RNA models of the 30S subunit A-site
Abstract
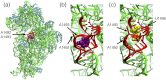
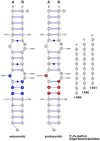
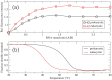
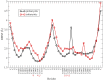
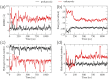
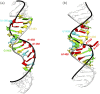
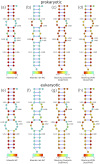
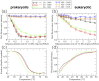
Synthetic oligonucleotides targeting functional regions of the prokaryotic rRNA could be promising antimicrobial agents. Indeed, such oligonucleotides were proven to inhibit bacterial growth. 2'-O-methylated (2'-O-Me) oligoribonucleotides with a sequence complementary to the decoding site in 16S rRNA were reported as inhibitors of bacterial translation. However, the binding mode and structures of the formed complexes, as well as the level of selectivity of the oligonucleotides between the prokaryotic and eukaryotic target, were not determined. We have analyzed three 2'-O-Me oligoribonucleotides designed to hybridize with the models of the prokaryotic rRNA containing two neighboring aminoglycoside binding pockets. One pocket is the paromomycin/kanamycin binding site corresponding to the decoding site in the small ribosomal subunit and the other one is the close-by hygromycin B binding site whose dynamics has not been previously reported. Molecular dynamics (MD) simulations, as well as isothermal titration calorimetry, gel electrophoresis and spectroscopic studies have shown that the eukaryotic rRNA model is less conformationally stable (in terms of hydrogen bonds and stacking interactions) than the corresponding prokaryotic one. In MD simulations of the eukaryotic construct, the nucleotide U1498, which plays an important role in correct positioning of mRNA during translation, is flexible and spontaneously flips out into the solvent. In solution studies, the 2'-O-Me oligoribonucleotides did not interact with the double stranded rRNA models but all formed stable complexes with the single-stranded prokaryotic target. 2'-O-Me oligoribonucleotides with one and two mismatches bound less tightly to the eukaryotic target. This shows that at least three mismatches between the 2'-O-Me oligoribonucleotide and eukaryotic rRNA are required to ensure target selectivity. The results also suggest that, in the ribosome environment, the strand invasion is the preferred binding mode of 2'-O-Me oligoribonucleotides targeting the aminoglycoside binding sites in 16S rRNA.
Conflict of interest statement
Figures



Similar articles
-
Targeting the A site RNA of the Escherichia coli ribosomal 30 S subunit by 2'-O-methyl oligoribonucleotides: a quantitative equilibrium dialysis binding assay and differential effects of aminoglycoside antibiotics.Biochem J. 2004 Oct 15;383(Pt 2):201-8. doi: 10.1042/BJ20040246. Biochem J. 2004. PMID: 15294017 Free PMC article.
-
Structural origins of aminoglycoside specificity for prokaryotic ribosomes.J Mol Biol. 2001 Mar 9;306(5):1037-58. doi: 10.1006/jmbi.2000.4420. J Mol Biol. 2001. PMID: 11237617
-
Analysis of ribosomal inter-subunit sites as targets for complementary oligonucleotides.Biopolymers. 2017 Apr;107(4). doi: 10.1002/bip.23004. Biopolymers. 2017. PMID: 27858985
-
Interactions of aminoglycoside antibiotics with rRNA.Biochem Soc Trans. 2016 Aug 15;44(4):987-93. doi: 10.1042/BST20160087. Biochem Soc Trans. 2016. PMID: 27528743 Review.
-
Antibiotic drugs targeting bacterial RNAs.Acta Pharm Sin B. 2014 Aug;4(4):258-65. doi: 10.1016/j.apsb.2014.06.012. Epub 2014 Jul 31. Acta Pharm Sin B. 2014. PMID: 26579393 Free PMC article. Review.
Cited by
-
Molecular Dynamics Study of the Hybridization between RNA and Modified Oligonucleotides.J Chem Theory Comput. 2019 Nov 12;15(11):6422-6432. doi: 10.1021/acs.jctc.9b00519. Epub 2019 Oct 9. J Chem Theory Comput. 2019. PMID: 31553600 Free PMC article.
-
Quantitative comparison between sub-millisecond time resolution single-molecule FRET measurements and 10-second molecular simulations of a biosensor protein.PLoS Comput Biol. 2020 Nov 5;16(11):e1008293. doi: 10.1371/journal.pcbi.1008293. eCollection 2020 Nov. PLoS Comput Biol. 2020. PMID: 33151943 Free PMC article.
-
Thermal Stability of Peptide Nucleic Acid Complexes.J Phys Chem B. 2019 Oct 3;123(39):8168-8177. doi: 10.1021/acs.jpcb.9b05168. Epub 2019 Sep 20. J Phys Chem B. 2019. PMID: 31491077 Free PMC article.
References
-
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484(7393):256–259. doi: 10.1038/nature10913 - DOI - PubMed
-
- Zeng X, Chugh J, Casiano-Negroni A, Al-Hashimi HM, Brooks CL. Flipping of the ribosomal A-site adenines provides a basis for tRNA selection. J Mol Biol. 2014;426(19):3201–3213. doi: 10.1016/j.jmb.2014.04.029 - DOI - PMC - PubMed
-
- Sanbonmatsu KY. Energy landscape of the ribosomal decoding center. Biochimie. 2006;88(8):1053–9. doi: 10.1016/j.biochi.2006.06.012 - DOI - PubMed
-
- Zhang W, Dunkle JA, Cate JHD. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325(5943):1014–7. doi: 10.1126/science.1175275 - DOI - PMC - PubMed
-
- Kietrys AM, Szopa A, Ba̧kowska-Żywicka K. Structure and function of intersubunit bridges in procaryotic ribosome. Biotechnologia. 2009;115(1):48–58.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

