Non-Coding RNAs As Transcriptional Regulators In Eukaryotes
- PMID: 29340213
- PMCID: PMC5762824
Non-Coding RNAs As Transcriptional Regulators In Eukaryotes
Abstract
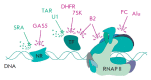
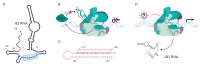
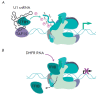
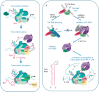
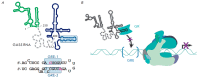
Non-coding RNAs up to 1,000 nucleotides in length are widespread in eukaryotes and fulfil various regulatory functions, in particular during chromatin remodeling and cell proliferation. These RNAs are not translated into proteins: thus, they are non-coding RNAs (ncRNAs). The present review describes the eukaryotic ncRNAs involved in transcription regulation, first and foremost, targeting RNA polymerase II (RNAP II) and/or its major proteinaceous transcription factors. The current state of knowledge concerning the regulatory functions of SRA and TAR RNA, 7SK and U1 snRNA, GAS5 and DHFR RNA is summarized herein. Special attention is given to murine B1 and B2 RNAs and human Alu RNA, due to their ability to bind the active site of RNAP II. Discovery of bacterial analogs of the eukaryotic small ncRNAs involved in transcription regulation, such as 6S RNAs, suggests that they possess a common evolutionary origin.
Keywords: RNA polymerase; noncoding RNAs; transcription regulation.
Figures
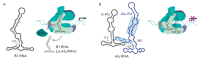
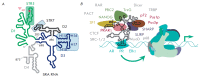
Similar articles
-
Small Noncoding 6S RNAs of Bacteria.Biochemistry (Mosc). 2015 Nov;80(11):1429-46. doi: 10.1134/S0006297915110048. Biochemistry (Mosc). 2015. PMID: 26615434 Review.
-
Non-coding RNAs regulating the transcriptional machinery.Biol Cell. 2008 Feb;100(2):83-95. doi: 10.1042/BC20070090. Biol Cell. 2008. PMID: 18199047 Review.
-
Noncoding RNAs Set the Stage for RNA Polymerase II Transcription.Trends Genet. 2021 Mar;37(3):279-291. doi: 10.1016/j.tig.2020.09.013. Epub 2020 Oct 9. Trends Genet. 2021. PMID: 33046273 Review.
-
TFIIF facilitates dissociation of RNA polymerase II from noncoding RNAs that lack a repression domain.Mol Cell Biol. 2010 Jan;30(1):91-7. doi: 10.1128/MCB.01115-09. Mol Cell Biol. 2010. PMID: 19841064 Free PMC article.
-
Regulation of transcription by 6S RNAs: insights from the Escherichia coli and Bacillus subtilis model systems.RNA Biol. 2014;11(5):508-21. doi: 10.4161/rna.28827. Epub 2014 Apr 23. RNA Biol. 2014. PMID: 24786589 Free PMC article. Review.
Cited by
-
The Role of microRNAs in Inflammation.Int J Mol Sci. 2022 Dec 7;23(24):15479. doi: 10.3390/ijms232415479. Int J Mol Sci. 2022. PMID: 36555120 Free PMC article. Review.
-
Regulatory role of long non coding RNAs (lncRNAs) in neurological disorders: From novel biomarkers to promising therapeutic strategies.Asian J Pharm Sci. 2021 Sep;16(5):533-550. doi: 10.1016/j.ajps.2021.02.006. Epub 2021 Apr 2. Asian J Pharm Sci. 2021. PMID: 34849161 Free PMC article. Review.
-
ALUminating the Path of Atherosclerosis Progression: Chaos Theory Suggests a Role for Alu Repeats in the Development of Atherosclerotic Vascular Disease.Int J Mol Sci. 2018 Jun 12;19(6):1734. doi: 10.3390/ijms19061734. Int J Mol Sci. 2018. PMID: 29895733 Free PMC article. Review.
-
Diversity and Versatility in Small RNA-Mediated Regulation in Bacterial Pathogens.Front Microbiol. 2021 Aug 10;12:719977. doi: 10.3389/fmicb.2021.719977. eCollection 2021. Front Microbiol. 2021. PMID: 34447363 Free PMC article. Review.
-
EZH2-interacting lncRNAs contribute to gastric tumorigenesis; a review on the mechanisms of action.Mol Biol Rep. 2024 Feb 23;51(1):334. doi: 10.1007/s11033-024-09237-7. Mol Biol Rep. 2024. PMID: 38393645 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
