Deep molecular phenotypes link complex disorders and physiological insult to CpG methylation
- PMID: 29325019
- PMCID: PMC5886112
- DOI: 10.1093/hmg/ddy006
Deep molecular phenotypes link complex disorders and physiological insult to CpG methylation
Abstract
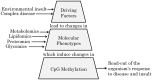
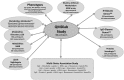
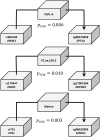
Epigenetic regulation of cellular function provides a mechanism for rapid organismal adaptation to changes in health, lifestyle and environment. Associations of cytosine-guanine di-nucleotide (CpG) methylation with clinical endpoints that overlap with metabolic phenotypes suggest a regulatory role for these CpG sites in the body's response to disease or environmental stress. We previously identified 20 CpG sites in an epigenome-wide association study (EWAS) with metabolomics that were also associated in recent EWASs with diabetes-, obesity-, and smoking-related endpoints. To elucidate the molecular pathways that connect these potentially regulatory CpG sites to the associated disease or lifestyle factors, we conducted a multi-omics association study including 2474 mass-spectrometry-based metabolites in plasma, urine and saliva, 225 NMR-based lipid and metabolite measures in blood, 1124 blood-circulating proteins using aptamer technology, 113 plasma protein N-glycans and 60 IgG-glyans, using 359 samples from the multi-ethnic Qatar Metabolomics Study on Diabetes (QMDiab). We report 138 multi-omics associations at these CpG sites, including diabetes biomarkers at the diabetes-associated TXNIP locus, and smoking-specific metabolites and proteins at multiple smoking-associated loci, including AHRR. Mendelian randomization suggests a causal effect of metabolite levels on methylation of obesity-associated CpG sites, i.e. of glycerophospholipid PC(O-36: 5), glycine and a very low-density lipoprotein (VLDL-A) on the methylation of the obesity-associated CpG loci DHCR24, MYO5C and CPT1A, respectively. Taken together, our study suggests that multi-omics-associated CpG methylation can provide functional read-outs for the underlying regulatory response mechanisms to disease or environmental insults.
Figures
Similar articles
-
Epigenome-wide association study (EWAS) on lipids: the Rotterdam Study.Clin Epigenetics. 2017 Feb 7;9:15. doi: 10.1186/s13148-016-0304-4. eCollection 2017. Clin Epigenetics. 2017. PMID: 28194238 Free PMC article.
-
DNA methylation and lipid metabolism: an EWAS of 226 metabolic measures.Clin Epigenetics. 2021 Jan 7;13(1):7. doi: 10.1186/s13148-020-00957-8. Clin Epigenetics. 2021. PMID: 33413638 Free PMC article.
-
Epigenome-wide association study on the plasma metabolome suggests self-regulation of the glycine and serine pathway through DNA methylation.Clin Epigenetics. 2024 Aug 13;16(1):104. doi: 10.1186/s13148-024-01718-7. Clin Epigenetics. 2024. PMID: 39138531 Free PMC article.
-
Blood-based epigenome-wide analyses of 19 common disease states: A longitudinal, population-based linked cohort study of 18,413 Scottish individuals.PLoS Med. 2023 Jul 6;20(7):e1004247. doi: 10.1371/journal.pmed.1004247. eCollection 2023 Jul. PLoS Med. 2023. PMID: 37410739 Free PMC article. Review.
-
Connecting the epigenome, metabolome and proteome for a deeper understanding of disease.J Intern Med. 2021 Sep;290(3):527-548. doi: 10.1111/joim.13306. Epub 2021 May 22. J Intern Med. 2021. PMID: 33904619 Review.
Cited by
-
Metabolite profiles and DNA methylation in metabolic syndrome: a two-sample, bidirectional Mendelian randomization.Front Genet. 2023 Sep 15;14:1184661. doi: 10.3389/fgene.2023.1184661. eCollection 2023. Front Genet. 2023. PMID: 37779905 Free PMC article.
-
AutoFocus: a hierarchical framework to explore multi-omic disease associations spanning multiple scales of biomolecular interaction.Commun Biol. 2024 Sep 6;7(1):1094. doi: 10.1038/s42003-024-06724-2. Commun Biol. 2024. PMID: 39237774 Free PMC article.
-
Structure Annotation of All Mass Spectra in Untargeted Metabolomics.Anal Chem. 2019 Feb 5;91(3):2155-2162. doi: 10.1021/acs.analchem.8b04698. Epub 2019 Jan 16. Anal Chem. 2019. PMID: 30608141 Free PMC article.
-
Epigenetics meets proteomics in an epigenome-wide association study with circulating blood plasma protein traits.Nat Commun. 2020 Jan 3;11(1):15. doi: 10.1038/s41467-019-13831-w. Nat Commun. 2020. PMID: 31900413 Free PMC article.
-
Elucidating the role of blood metabolites on pancreatic cancer risk using two-sample Mendelian randomization analysis.Int J Cancer. 2024 Mar 1;154(5):852-862. doi: 10.1002/ijc.34771. Epub 2023 Oct 20. Int J Cancer. 2024. PMID: 37860916
References
-
- Banovich N.E., Lan X., McVicker G., van de Geijn B., Degner J.F., Blischak J.D., Roux J., Pritchard J.K., Gilad Y.. et al. (2014) Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genetics, 10, e1004663.. - PMC - PubMed
-
- Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., Heine-Suner D., Cigudosa J.C., Urioste M., Benitez J.. et al. (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America, 102, 10604–10609. - PMC - PubMed
-
- Chambers J.C., Loh M., Lehne B., Drong A., Kriebel J., Motta V., Wahl S., Elliott H.R., Rota F., Scott W.R.. et al. (2015) Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. The Lancet Diabetes & Endocrinology, 3, 526–534. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

