Caveolin-mediated endocytosis of the Chlamydia M278 outer membrane peptide encapsulated in poly(lactic acid)-Poly(ethylene glycol) nanoparticles by mouse primary dendritic cells enhances specific immune effectors mediated by MHC class II and CD4+ T cells
- PMID: 29324305
- PMCID: PMC5801148
- DOI: 10.1016/j.biomaterials.2017.12.019
Caveolin-mediated endocytosis of the Chlamydia M278 outer membrane peptide encapsulated in poly(lactic acid)-Poly(ethylene glycol) nanoparticles by mouse primary dendritic cells enhances specific immune effectors mediated by MHC class II and CD4+ T cells
Abstract
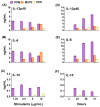
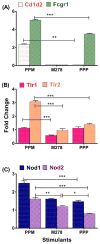
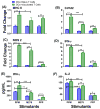
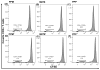
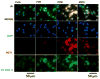
We previously developed a Chlamydia trachomatis nanovaccine (PPM) by encapsulating a chlamydial M278 peptide within poly(lactic acid)-poly(ethylene glycol) biodegradable nanoparticles that immunopotentiated Chlamydia-specific immune effector responses in mice. Herein, we investigated the mechanistic interactions of PPM with mouse bone marrow-derived dendritic cells (DCs) for its uptake, trafficking, and T cell activation. Our results reveal that PPM triggered enhanced expression of effector cytokines and chemokines, surface activation markers (Cd1d2, Fcgr1), pathogen-sensing receptors (TLR2, Nod1), co-stimulatory (CD40, CD80, CD86) and MHC class I and II molecules. Co-culturing of PPM-primed DCs with T cells from C. muridarum vaccinated mice yielded an increase in Chlamydia-specific immune effector responses including CD3+ lymphoproliferation, CD3+CD4+ IFN-γ-secreting cells along with CD3+CD4+ memory (CD44high and CD62Lhigh) and effector (CD44high and CD62Llow) phenotypes. Intracellular trafficking analyses revealed an intense expression and colocalization of PPM predominantly in endosomes. PPM also upregulated the transcriptional and protein expression of the endocytic mediator, caveolin-1 in DCs. More importantly, the specific inhibition of caveolin-1 led to decreased expression of PPM-induced cytokines and co-stimulatory molecules. Our investigation shows that PPM provided enhancement of uptake, probably by exploiting the caveolin-mediated endocytosis pathway, endosomal processing, and MHC II presentation to immunopotentiate Chlamydia-specific immune effector responses mediated by CD4+ T cells.
Keywords: Caveolin; Chlamydia muridarum; Dendritic cells; Endocytosis; Nanovaccine; PLA-PEG nanoparticles.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures










Similar articles
-
The Chlamydia M278 Major Outer Membrane Peptide Encapsulated in the Poly(lactic acid)-Poly(ethylene glycol) Nanoparticulate Self-Adjuvanting Delivery System Protects Mice Against a Chlamydia muridarum Genital Tract Challenge by Stimulating Robust Systemic and Local Mucosal Immune Responses.Front Immunol. 2018 Oct 15;9:2369. doi: 10.3389/fimmu.2018.02369. eCollection 2018. Front Immunol. 2018. PMID: 30374357 Free PMC article.
-
Poly(lactic acid)-poly(ethylene glycol) nanoparticles provide sustained delivery of a Chlamydia trachomatis recombinant MOMP peptide and potentiate systemic adaptive immune responses in mice.Nanomedicine. 2014 Aug;10(6):1311-21. doi: 10.1016/j.nano.2014.02.009. Epub 2014 Mar 4. Nanomedicine. 2014. PMID: 24602605 Free PMC article.
-
A nanovaccine formulation of Chlamydia recombinant MOMP encapsulated in PLGA 85:15 nanoparticles augments CD4+ effector (CD44high CD62Llow) and memory (CD44high CD62Lhigh) T-cells in immunized mice.Nanomedicine. 2020 Oct;29:102257. doi: 10.1016/j.nano.2020.102257. Epub 2020 Jun 28. Nanomedicine. 2020. PMID: 32610072 Free PMC article.
-
Outer membrane proteins preferentially load MHC class II peptides: implications for a Chlamydia trachomatis T cell vaccine.Vaccine. 2015 Apr 27;33(18):2159-66. doi: 10.1016/j.vaccine.2015.02.055. Epub 2015 Mar 1. Vaccine. 2015. PMID: 25738816 Free PMC article.
-
Encapsulation of Recombinant MOMP in Extended-Releasing PLGA 85:15 Nanoparticles Confer Protective Immunity Against a Chlamydia muridarum Genital Challenge and Re-Challenge.Front Immunol. 2021 Apr 14;12:660932. doi: 10.3389/fimmu.2021.660932. eCollection 2021. Front Immunol. 2021. PMID: 33936096 Free PMC article.
Cited by
-
Effects of prime-boost strategies on the protective efficacy and immunogenicity of a PLGA (85:15)-encapsulated Chlamydia recombinant MOMP nanovaccine.Pathog Dis. 2024 Feb 7;82:ftae004. doi: 10.1093/femspd/ftae004. Pathog Dis. 2024. PMID: 38862192 Free PMC article.
-
PLGA-Chitosan Encapsulated IL-10 Nanoparticles Modulate Chlamydia Inflammation in Mice.Int J Nanomedicine. 2024 Feb 8;19:1287-1301. doi: 10.2147/IJN.S432970. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38348174 Free PMC article.
-
A new temperature-dependent strategy to modulate the epidermal growth factor receptor.Biomaterials. 2018 Nov;183:319-330. doi: 10.1016/j.biomaterials.2018.07.063. Epub 2018 Aug 11. Biomaterials. 2018. PMID: 30196151 Free PMC article.
-
Irradiation pretreatment enhances the therapeutic efficacy of platelet-membrane-camouflaged antitumor nanoparticles.J Nanobiotechnology. 2020 Jul 20;18(1):101. doi: 10.1186/s12951-020-00660-z. J Nanobiotechnology. 2020. PMID: 32690018 Free PMC article.
-
Prolonged Release and Functionality of Interleukin-10 Encapsulated within PLA-PEG Nanoparticles.Nanomaterials (Basel). 2019 Jul 26;9(8):1074. doi: 10.3390/nano9081074. Nanomaterials (Basel). 2019. PMID: 31357440 Free PMC article.
References
-
- Kalkanidis M, Pietersz GA, Xiang SD, Mottram PL, Crimeen-Irwin B, Ardipradja K, et al. Methods for nano-particle based vaccine formulation and evaluation of their immunogenicity. Methods. 2006;40:20–9. - PubMed
-
- Bachmann MF, Jennings GT. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10:787–96. - PubMed
-
- Jennings GT, Bachmann MF. Designing recombinant vaccines with viral properties: a rational approach to more effective vaccines. Curr Mol Med. 2007;7:143–55. - PubMed
-
- Shi Y, Huang G. Recent developments of biodegradable and biocompatible materials based micro/nanoparticles for delivering macromolecular therapeutics. Crit Rev Ther Drug Carrier Syst. 2009;26:29–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

