Impact of protein-ligand solvation and desolvation on transition state thermodynamic properties of adenosine A2A ligand binding kinetics
- PMID: 29308352
- PMCID: PMC5755719
- DOI: 10.1007/s40203-017-0037-x
Impact of protein-ligand solvation and desolvation on transition state thermodynamic properties of adenosine A2A ligand binding kinetics
Abstract
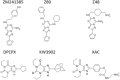
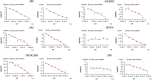
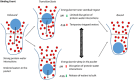
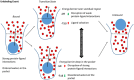
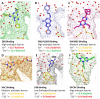
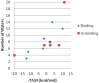
Ligand-protein binding kinetic rates are growing in importance as parameters to consider in drug discovery and lead optimization. In this study we analysed using surface plasmon resonance (SPR) the transition state (TS) properties of a set of six adenosine A2A receptor inhibitors, belonging to both the xanthine and the triazolo-triazine scaffolds. SPR highlighted interesting differences among the ligands in the enthalpic and entropic components of the TS energy barriers for the binding and unbinding events. To better understand at a molecular level these differences, we developed suMetaD, a novel molecular dynamics (MD)-based approach combining supervised MD and metadynamics. This method allows simulation of the ligand unbinding and binding events. It also provides the system conformation corresponding to the highest energy barrier the ligand is required to overcome to reach the final state. For the six ligands evaluated in this study their TS thermodynamic properties were linked in particular to the role of water molecules in solvating/desolvating the pocket and the small molecules. suMetaD identified kinetic bottleneck conformations near the bound state position or in the vestibule area. In the first case the barrier is mainly enthalpic, requiring the breaking of strong interactions with the protein. In the vestibule TS location the kinetic bottleneck is instead mainly of entropic nature, linked to the solvent behaviour.
Keywords: Biacore; Ligand binding kinetics; Metadynamics; Molecular dynamics; SPR; Supervised molecular dynamics.
Figures

Similar articles
-
Combined Free-Energy Calculation and Machine Learning Methods for Understanding Ligand Unbinding Kinetics.J Chem Theory Comput. 2022 Apr 12;18(4):2543-2555. doi: 10.1021/acs.jctc.1c00924. Epub 2022 Feb 23. J Chem Theory Comput. 2022. PMID: 35195418 Free PMC article.
-
Decoding the Role of Water Dynamics in Ligand-Protein Unbinding: CRF1R as a Test Case.J Chem Inf Model. 2015 Sep 28;55(9):1857-66. doi: 10.1021/acs.jcim.5b00440. Epub 2015 Sep 10. J Chem Inf Model. 2015. PMID: 26335976
-
Contributions of water transfer energy to protein-ligand association and dissociation barriers: Watermap analysis of a series of p38α MAP kinase inhibitors.Proteins. 2013 Sep;81(9):1509-26. doi: 10.1002/prot.24276. Epub 2013 Jul 2. Proteins. 2013. PMID: 23468227
-
Surface plasmon resonance spectroscopy for characterisation of membrane protein-ligand interactions and its potential for drug discovery.Biochim Biophys Acta. 2014 Jan;1838(1 Pt A):43-55. doi: 10.1016/j.bbamem.2013.04.028. Epub 2013 May 9. Biochim Biophys Acta. 2014. PMID: 23665295 Review.
-
Estimation of kinetic and thermodynamic ligand-binding parameters using computational strategies.Future Med Chem. 2017 Apr;9(5):507-523. doi: 10.4155/fmc-2016-0224. Epub 2017 Mar 31. Future Med Chem. 2017. PMID: 28362130 Review.
Cited by
-
Metadynamics simulations leveraged by statistical analyses and artificial intelligence-based tools to inform the discovery of G protein-coupled receptor ligands.Front Endocrinol (Lausanne). 2022 Dec 23;13:1099715. doi: 10.3389/fendo.2022.1099715. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36619585 Free PMC article. Review.
-
In Silico Drug Design for Purinergic GPCRs: Overview on Molecular Dynamics Applied to Adenosine and P2Y Receptors.Biomolecules. 2020 May 26;10(6):812. doi: 10.3390/biom10060812. Biomolecules. 2020. PMID: 32466404 Free PMC article. Review.
-
Selective activation of Gαob by an adenosine A1 receptor agonist elicits analgesia without cardiorespiratory depression.Nat Commun. 2022 Jul 18;13(1):4150. doi: 10.1038/s41467-022-31652-2. Nat Commun. 2022. PMID: 35851064 Free PMC article.
-
Membrane Lipids Are an Integral Part of Transmembrane Allosteric Sites in GPCRs: A Case Study of Cannabinoid CB1 Receptor Bound to a Negative Allosteric Modulator, ORG27569, and Analogs.J Med Chem. 2022 Sep 22;65(18):12240-12255. doi: 10.1021/acs.jmedchem.2c00946. Epub 2022 Sep 6. J Med Chem. 2022. PMID: 36066412 Free PMC article.
-
New Insights into Key Determinants for Adenosine 1 Receptor Antagonists Selectivity Using Supervised Molecular Dynamics Simulations.Biomolecules. 2020 May 7;10(5):732. doi: 10.3390/biom10050732. Biomolecules. 2020. PMID: 32392873 Free PMC article.
References
-
- Barducci A, Bonomi M, Parrinello M. Metadynamics: metadynamics. Wiley Interdiscip Rev Comput Mol Sci. 2011;1:826–843. doi: 10.1002/wcms.31. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

