Robo1 Forms a Compact Dimer-of-Dimers Assembly
- PMID: 29307485
- PMCID: PMC5807052
- DOI: 10.1016/j.str.2017.12.003
Robo1 Forms a Compact Dimer-of-Dimers Assembly
Abstract
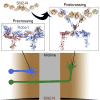
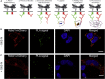
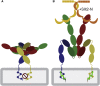
Roundabout (Robo) receptors provide an essential repulsive cue in neuronal development following Slit ligand binding. This important signaling pathway can also be hijacked in numerous cancers, making Slit-Robo an attractive therapeutic target. However, little is known about how Slit binding mediates Robo activation. Here we present the crystal structure of Robo1 Ig1-4 and Robo1 Ig5, together with a negative stain electron microscopy reconstruction of the Robo1 ectodomain. These results show how the Robo1 ectodomain is arranged as compact dimers, mainly mediated by the central Ig domains, which can further interact in a "back-to-back" fashion to generate a tetrameric assembly. We also observed no change in Robo1 oligomerization upon interaction with the dimeric Slit2-N ligand using fluorescent imaging. Taken together with previous studies we propose that Slit2-N binding results in a conformational change of Robo1 to trigger cell signaling.
Keywords: X-ray crystallography; electron microscopy; receptor; robo; slit.
Copyright © 2017 European Molecular Biology Laboratory. Published by Elsevier Ltd.. All rights reserved.
Figures
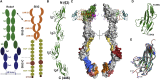

Comment in
-
Approaching the Roundabout: cis and trans Robo1 Contacts Revealed.Structure. 2018 Feb 6;26(2):183-184. doi: 10.1016/j.str.2018.01.012. Structure. 2018. PMID: 29413320
Similar articles
-
Minimal structural elements required for midline repulsive signaling and regulation of Drosophila Robo1.PLoS One. 2020 Oct 22;15(10):e0241150. doi: 10.1371/journal.pone.0241150. eCollection 2020. PLoS One. 2020. PMID: 33091076 Free PMC article.
-
Slit Binding via the Ig1 Domain Is Essential for Midline Repulsion by Drosophila Robo1 but Dispensable for Receptor Expression, Localization, and Regulation in Vivo.G3 (Bethesda). 2015 Sep 10;5(11):2429-39. doi: 10.1534/g3.115.022327. G3 (Bethesda). 2015. PMID: 26362767 Free PMC article.
-
In vivo functional analysis of Drosophila Robo1 immunoglobulin-like domains.Neural Dev. 2016 Aug 18;11(1):15. doi: 10.1186/s13064-016-0071-0. Neural Dev. 2016. PMID: 27539083 Free PMC article.
-
Slit-Robo signaling.Development. 2016 Sep 1;143(17):3037-44. doi: 10.1242/dev.132829. Development. 2016. PMID: 27578174 Review.
-
Structure and Function of Roundabout Receptors.Subcell Biochem. 2019;93:291-319. doi: 10.1007/978-3-030-28151-9_9. Subcell Biochem. 2019. PMID: 31939155 Review.
Cited by
-
Robo functions as an attractive cue for glial migration through SYG-1/Neph.Elife. 2020 Nov 19;9:e57921. doi: 10.7554/eLife.57921. Elife. 2020. PMID: 33211005 Free PMC article.
-
Generation of biparatopic antibody through two-step targeting of fragment antibodies on antigen using SpyTag and SpyCatcher.Biotechnol Rep (Amst). 2020 Jan 11;25:e00418. doi: 10.1016/j.btre.2020.e00418. eCollection 2020 Mar. Biotechnol Rep (Amst). 2020. PMID: 31993343 Free PMC article.
-
Cell guidance ligands, receptors and complexes - orchestrating signalling in time and space.Curr Opin Struct Biol. 2020 Apr;61:79-85. doi: 10.1016/j.sbi.2019.11.007. Epub 2019 Dec 17. Curr Opin Struct Biol. 2020. PMID: 31862615 Free PMC article. Review.
-
Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility.Structure. 2020 May 5;28(5):507-515.e5. doi: 10.1016/j.str.2020.02.008. Epub 2020 Mar 17. Structure. 2020. PMID: 32187531 Free PMC article.
-
NMR analysis suggests the terminal domains of Robo1 remain extended but are rigidified in the presence of heparan sulfate.Sci Rep. 2022 Aug 30;12(1):14769. doi: 10.1038/s41598-022-18769-6. Sci Rep. 2022. PMID: 36042257 Free PMC article.
References
-
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. - PubMed
-
- Barak R., Lahmi R., Gevorkyan-Airapetov L., Levy E., Tzur A., Opatowsky Y. Crystal structure of the extracellular juxtamembrane region of Robo1. J. Struct. Biol. 2014;186:283–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

