Development and Evaluation of a Broad Bead-Based Multiplex Immunoassay To Measure IgG Seroreactivity against Human Polyomaviruses
- PMID: 29305551
- PMCID: PMC5869810
- DOI: 10.1128/JCM.01566-17
Development and Evaluation of a Broad Bead-Based Multiplex Immunoassay To Measure IgG Seroreactivity against Human Polyomaviruses
Abstract
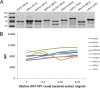
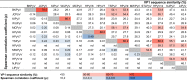
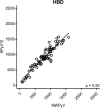
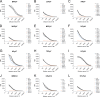
The family of polyomaviruses, which cause severe disease in immunocompromised hosts, has expanded substantially in recent years. To accommodate measurement of IgG seroresponses against all currently known human polyomaviruses (HPyVs), including the Lyon IARC polyomavirus (LIPyV), we extended our custom multiplex bead-based HPyV immunoassay and evaluated the performance of this pan-HPyV immunoassay. The VP1 proteins of 15 HPyVs belonging to 13 Polyomavirus species were expressed as recombinant glutathione S-transferase (GST) fusion proteins and coupled to fluorescent Luminex beads. Sera from healthy blood donors and immunocompromised kidney transplant recipients were used to analyze seroreactivity against the different HPyVs. For BK polyomavirus (BKPyV), the GST-VP1 fusion protein-directed seroresponses were compared to those obtained against BKPyV VP1 virus-like particles (VLP). Seroreactivity against most HPyVs was common and generally high in both test populations. Low seroreactivity against HPyV9, HPyV12, New Jersey PyV, and LIPyV was observed. The assay was reproducible (Pearson's r2 > 0.84, P < 0.001) and specific. Weak but consistent cross-reactivity between the related viruses HPyV6 and HPyV7 was observed. The seroresponses measured by the GST-VP1-based immunoassay and a VP1 VLP-based enzyme-linked immunosorbent assay were highly correlated (Spearman's ρ = 0.823, P < 0.001). The bead-based pan-HPyV multiplex immunoassay is a reliable tool to determine HPyV-specific seroresponses with high reproducibility and specificity and is suitable for use in seroepidemiological studies.
Keywords: immunoassay; immunology; polyomavirus; seroepidemiology.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV and KIPyV polyomaviruses among the healthy blood donors.J Med Virol. 2016 Jul;88(7):1254-61. doi: 10.1002/jmv.24440. Epub 2015 Dec 15. J Med Virol. 2016. PMID: 26630080
-
Seroprevalence of fourteen human polyomaviruses determined in blood donors.PLoS One. 2018 Oct 23;13(10):e0206273. doi: 10.1371/journal.pone.0206273. eCollection 2018. PLoS One. 2018. PMID: 30352098 Free PMC article.
-
Production of recombinant VP1-derived virus-like particles from novel human polyomaviruses in yeast.BMC Biotechnol. 2015 Aug 4;15:68. doi: 10.1186/s12896-015-0187-z. BMC Biotechnol. 2015. PMID: 26239840 Free PMC article.
-
Serological cross-reactivity between human polyomaviruses.Rev Med Virol. 2013 Jul;23(4):250-64. doi: 10.1002/rmv.1747. Epub 2013 May 6. Rev Med Virol. 2013. PMID: 23650080 Review.
-
[New, newer, newest human polyomaviruses: how far?].Mikrobiyol Bul. 2013 Apr;47(2):362-81. doi: 10.5578/mb.5377. Mikrobiyol Bul. 2013. PMID: 23621738 Review. Turkish.
Cited by
-
Prevalence of DNA of fourteen human polyomaviruses determined in blood donors.Transfusion. 2019 Dec;59(12):3689-3697. doi: 10.1111/trf.15557. Epub 2019 Oct 21. Transfusion. 2019. PMID: 31633816 Free PMC article.
-
Novel polyomaviruses identified in fecal samples from four carnivore species.Arch Virol. 2023 Jan 3;168(1):18. doi: 10.1007/s00705-022-05675-5. Arch Virol. 2023. PMID: 36593361 Free PMC article.
-
Tetraplex Fluorescent Microbead-Based Immunoassay for the Serodiagnosis of Newcastle Disease Virus and Avian Influenza Viruses in Poultry Sera.Pathogens. 2022 Sep 17;11(9):1059. doi: 10.3390/pathogens11091059. Pathogens. 2022. PMID: 36145491 Free PMC article.
-
Prevalence and viral loads of polyomaviruses BKPyV, JCPyV, MCPyV, TSPyV and NJPyV and hepatitis viruses HBV, HCV and HEV in HIV-infected patients in China.Sci Rep. 2020 Oct 13;10(1):17066. doi: 10.1038/s41598-020-74244-0. Sci Rep. 2020. PMID: 33051567 Free PMC article.
-
Immune Determinants of Viral Clearance in Hospitalised COVID-19 Patients: Reduced Circulating Naïve CD4+ T Cell Counts Correspond with Delayed Viral Clearance.Cells. 2022 Sep 2;11(17):2743. doi: 10.3390/cells11172743. Cells. 2022. PMID: 36078151 Free PMC article.
References
-
- Gheit T, Dutta S, Oliver J, Robitaille A, Hampras S, Combes J-D, McKay-Chopin S, Le Calvez-Kelm F, Fenske N, Cherpelis B, Giuliano AR, Franceschi S, McKay J, Rollison DE, Tommasino M. 2017. Isolation and characterization of a novel putative human polyomavirus. Virology 506:45–54. doi:10.1016/j.virol.2017.03.007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

