Engineering posttranslational proofreading to discriminate nonstandard amino acids
- PMID: 29301968
- PMCID: PMC5776986
- DOI: 10.1073/pnas.1715137115
Engineering posttranslational proofreading to discriminate nonstandard amino acids
Abstract
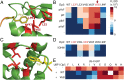
Incorporation of nonstandard amino acids (nsAAs) leads to chemical diversification of proteins, which is an important tool for the investigation and engineering of biological processes. However, the aminoacyl-tRNA synthetases crucial for this process are polyspecific in regard to nsAAs and standard amino acids. Here, we develop a quality control system called "posttranslational proofreading" to more accurately and rapidly evaluate nsAA incorporation. We achieve this proofreading by hijacking a natural pathway of protein degradation known as the N-end rule, which regulates the lifespan of a protein based on its amino-terminal residue. We find that proteins containing certain desired N-terminal nsAAs have much longer half-lives compared with those proteins containing undesired amino acids. We use the posttranslational proofreading system to further evolve a Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (TyrRS) variant and a tRNATyr species for improved specificity of the nsAA biphenylalanine in vitro and in vivo. Our newly evolved biphenylalanine incorporation machinery enhances the biocontainment and growth of genetically engineered Escherichia coli strains that depend on biphenylalanine incorporation. Finally, we show that our posttranslational proofreading system can be designed for incorporation of other nsAAs by rational engineering of the ClpS protein, which mediates the N-end rule. Taken together, our posttranslational proofreading system for in vivo protein sequence verification presents an alternative paradigm for molecular recognition of amino acids and is a major advance in our ability to accurately expand the genetic code.
Keywords: N-end rule; genetic code expansion; nonstandard amino acids; protein degradation; synthetic biology.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: G.M.C. has related financial interests in ReadCoor, EnEvolv, and GRO Biosciences. A.M.K. and G.M.C. have filed a provisional patent on posttranslational proofreading, and A.M.K., D.S., E.K., and G.M.C. have filed a provisional patent on evolved BipA OTS variants. For a complete list of G.M.C.’s financial interests, please visit arep.med.harvard.edu/gmc/tech.html.
Figures


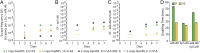
Similar articles
-
Improved Incorporation of Noncanonical Amino Acids by an Engineered tRNA(Tyr) Suppressor.Biochemistry. 2016 Jan 26;55(3):618-28. doi: 10.1021/acs.biochem.5b01185. Epub 2016 Jan 8. Biochemistry. 2016. PMID: 26694948 Free PMC article.
-
Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion.Nucleic Acids Res. 2010 Jun;38(11):3682-91. doi: 10.1093/nar/gkq080. Epub 2010 Feb 16. Nucleic Acids Res. 2010. PMID: 20159998 Free PMC article.
-
Selective and Site-Specific Incorporation of Nonstandard Amino Acids Within Proteins for Therapeutic Applications.Methods Mol Biol. 2024;2720:35-53. doi: 10.1007/978-1-0716-3469-1_3. Methods Mol Biol. 2024. PMID: 37775656
-
Semisynthetic Organisms with Expanded Genetic Codes.Biochemistry. 2018 Apr 17;57(15):2177-2178. doi: 10.1021/acs.biochem.8b00013. Epub 2018 Apr 6. Biochemistry. 2018. PMID: 29624384 Free PMC article. Review.
-
Incorporation of non-standard amino acids into proteins: challenges, recent achievements, and emerging applications.Appl Microbiol Biotechnol. 2019 Apr;103(7):2947-2958. doi: 10.1007/s00253-019-09690-6. Epub 2019 Feb 21. Appl Microbiol Biotechnol. 2019. PMID: 30790000 Free PMC article. Review.
Cited by
-
Advances in engineering microbial biosynthesis of aromatic compounds and related compounds.Bioresour Bioprocess. 2021 Sep 27;8(1):91. doi: 10.1186/s40643-021-00434-x. Bioresour Bioprocess. 2021. PMID: 38650203 Free PMC article. Review.
-
Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation.Genes (Basel). 2018 Dec 28;10(1):17. doi: 10.3390/genes10010017. Genes (Basel). 2018. PMID: 30597824 Free PMC article. Review.
-
A swapped genetic code prevents viral infections and gene transfer.Nature. 2023 Mar;615(7953):720-727. doi: 10.1038/s41586-023-05824-z. Epub 2023 Mar 15. Nature. 2023. PMID: 36922599 Free PMC article.
-
The expanded specificity and physiological role of a widespread N-degron recognin.Proc Natl Acad Sci U S A. 2019 Sep 10;116(37):18629-18637. doi: 10.1073/pnas.1821060116. Epub 2019 Aug 26. Proc Natl Acad Sci U S A. 2019. PMID: 31451664 Free PMC article.
-
A platform for distributed production of synthetic nitrated proteins in live bacteria.Nat Chem Biol. 2023 Jul;19(7):911-920. doi: 10.1038/s41589-023-01338-x. Epub 2023 May 15. Nat Chem Biol. 2023. PMID: 37188959
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

